Journal of
eISSN: 2373-437X


Research Article Volume 5 Issue 1
1Ethiopian Institute of Agricultural Research, Ethiopia
2Federal University of Vicosa, Brazil
3Embrapa Coffee Genetics and Improvement Researcher, Federal University of Vicosa, Brazil
Correspondence: Bayisa Asefa Bikila, Ethiopian Institute of Agricultural Research, Kulumsa Research Center, PO Box 489, Asella, Ethiopia
Received: February 23, 2017 | Published: May 19, 2017
Citation: Bikila BA, Sakiyama NS, Caixeta ET (2017) SNPs Based Molecular Diversity of Coffea canephora. J Microbiol Exp 5(1): 00136. DOI: 10.15406/jmen.2017.05.00136
Coffee offers one of the most widely drunk beverages in the world, and is a very important source of foreign currency income for many countries. Coffea canephora Pierre presents a great genetic variability, with one of the widest natural geographical distribution within the subgenus. Newly developed single nucleotide Polymorphism (SNP) markers are effective in genetic diversity detection. The objective of this study was to analyze the genetic diversity and group C. canephora accessions, which were genotyped with SNPs molecular markers. In the present study, C. canephora germplasm consisting of 50 clones (24 Conilon and 26 Robusta) were used. Genetic diversity was investigated using 46074 polymorphic SNP markers covering the entire genome of C. canephora. The estimation of genetic similarity between each pair of individuals was calculated by the Jaccard coefficient using the program NTSYS pc2.1. A simplified representation of the similarity was obtained by constructing the dendrogram using UPGMA method (Unweighted Pair-Group Method using Arithmetic Mean). The optimal number of groups in the dendrogram was determined by the relative size of distances in the dendrogram. Thus, the first group was composed of clones 1, 5, 24, 6, 9, 14, 23, 22, 10, 17, 19, 18, 20, 2, 13, 11, 15, 7, 4 , 3 and 21; the second group 8, the third group 12, the fourth 16, the fifth group 26, 31, 33, 34, 37, 38, 28, 45, 46, 41 25, 29, 36, 43, 48, 49, 47, 50, 35, 39, 44, 40, 32, 30, and the sixth composed by 27 and 42. Generally, the analysis showed that the C. canephora clones were clearly divided into diversity groups that can be used for further breeding programs.
Keywords: Genetic diversity; Genetic similarity; Molecular markers
Coffee provides one of the most widely drunk beverages in the world, and is a very important source of foreign exchange income for many countries, and ranks second on international trade exchanges, representing a significant source of income to several developing countries in Africa, Asia and Latin America. Coffee belongs to the Family, Rubiaceae. The genus Coffea L. comprises 104 taxa that have been identified so far. Commercial coffee production relies mainly on two species, Coffea arabica L. (63%) and Coffea canephora Pierre (36%). C. arabica is a natural allotetraploid (2n = 4X = 44), and is self-fertile.1. Other species are diploids (2n = 2x= 22) and generally self-incompatible. Unlike C. arabica, plants of C. canephora does not need to grow at high altitude, requires less care to grow because it is hardier and it tends to be less susceptible to pests and rough handling.1 Its area of distribution is variable and corresponds to hot and humid climatic regions.
Globally, the C. canephora gene pool is conserved in ex-situ collection plots in several countries. In cognizant of this fact and in order to alleviate the production problems, concerted effort were undertaken to utilize coffea canephora germplasm, as result several cultivars and accessions were collected and maintained at the different centers in Brazil. The main coffee germplasm collections are placed in governmental institutions, in São Paulo, at Instituto Agronômico de Campinas (IAC); in Espírito Santo, at Instituto Capixaba de Pesquisa, Assistência Técnica e Extensão Rural (Incaper); in Minas Gerais, at the Universidade Federal de Viçosa (UFV), in a partnership with Empresa de Pesquisa Agropecuária de Minas Gerais (Epamig); and in Rondônia, at Embrapa-Rondônia. Furthermore, Espírito Santo and Rondônia response for 75% of C. canephora produced in Brazil. Embrapa-Rondônia has the particularity of containing a significant number of accessions resulting from exchanges with the institutions mentioned before (i.e.: IAC, UFV and Incaper), besides an expressive sample of local genotypes.2 Consequently, its variability is representative of the germplasm commercially grown and conserved in Brazil.
C. canephora Pierre presents a wide genetic variability, with one of the widest geographic natural distribution within the subgenus Coffea.3 In view of this, characterization and evaluation of its gene pool is necessary for effective crop improvement programmes and for better conservation and management of genetic resources.4 Assessment of genetic variability within and among arabica coffee populations using molecular markers such as RAPD, ISSR, ALFP, and SSR has been the subject of several studies.5-9 Similar analysis in C. canephorahas paramount importance, especially for genetic materials from Brazil.
With the advent of SNP (Single Nucleotide Polymorphism) markers, the possibility of simultaneous analysis of a set of loci becomes more real. A SNP is created when a single nucleotide base in a DNA sequence is replaced with a different nucleotide base. The SNP markers are based on the most fundamental alterations of the DNA molecule, mutations in the bases of unique chain of nitrogenous bases (Adenine, Cytosine, Guanine and Thymine). The SNP are extremely abundant in genomes, studies show that there may be millions in an individual genome.10
The genetic variation of a quantitative trait is controlled by segregation of multiple genes described by the infinitesimal model of Fisher,11 which assumes that the number of loci is infinitely large and each with small effect. The genetic variances of individual loci are so small that they cannot be investigated individually and necessary to analysis sets of these loci. Thus, the availability of dense marker maps has opened new opportunities for genetic evaluation of individuals with high accuracy. Hence, this study was designed to estimate the level of genetic diversity and relationship among and within the clones of C. canephora conserved at research station of UFV using SNPs markers which can capture the whole genetic variability and useful for future coffee improvement program.
Plant materials
A total of 50 C. canephora genotypes including two varietal groups, 26 Robusta and 24 Conilon, maintained at the Coffee Germplasm Collection of UFV, Minas Gerais, Brazil, were used for this study (Table 1).
No. |
Conilon (UFV Code) |
No. |
Robusta (UFV Code) |
1 |
UFV513 |
25 |
UFV 3365-144 |
2 |
UFV 3627-31 |
26 |
UFV 3366-139 |
3 |
UFV 3628-2 |
27 |
UFV3373-36 |
4 |
UFV 3629-11 |
28 |
UFV3374-28 |
5 |
UFV 3629-25 |
29 |
UFV514 |
6 |
UFV3627-20 |
30 |
UFV3356-71 |
7 |
UFV3627-24 |
31 |
UFV3357-93 |
8 |
UFV 3627-27 |
32 |
UFV3358-88 |
9 |
UFV 3627-29 |
33 |
UFV3360-169 |
10 |
UFV 3627-30 |
34 |
UFV3361-148 |
11 |
UFV 3628-1 |
35 |
UFV3363-118 |
12 |
UFV 3628-3 |
36 |
UFV3366-134 |
13 |
UFV 3628-5 |
37 |
UFV3367-101 |
14 |
UFV 3628-16 |
38 |
UFV3368-58 |
15 |
UFV 3628-29 |
39 |
UFV3370-47 |
16 |
UFV 3628-37 |
40 |
UFV3371-19 |
17 |
UFV 3628-45 |
41 |
UFV3373-43 |
18 |
UFV 3629-4 |
42 |
UFV3374-29 |
19 |
UFV 3629-7 |
43 |
UFV3375-65 |
20 |
UFV 3629-10 |
44 |
UFV3376-8 |
21 |
UFV 3629-17 |
45 |
UFV3630-2 |
22 |
UFV 3629-27 |
46 |
UFV3631-1 |
23 |
UFV 3629-29 |
47 |
UFV 3631-6 |
24 |
UFV 3629-30 |
48 |
UFV3356-74 |
49 |
UFV3631-9 |
||
|
|
50 |
UFV3631-13 |
Table 1 List of Coffea canephora clones used for genetic diversity study
Genotyping with SNP markers
For DNA extraction, young and completely extended leaves were collected from each accession, frozen at -80 °C, lyophilized, ground to make a fine powder and kept at -20 °C until used. Genomic DNA was extracted using the method described by Diniz et al..12 SNPs were genotyped in Florida, USA using the RAPiD-Seq technology platform.
Statistical Analysis
The genetic similarity (Sgij), between pairs of C. canephora individuals was calculated using the Jaccard coefficient. The similarities were calculated employing 4607 SNPs markers, using the following expression:

Sgij where:
a = Number of cases in which the presence of the band occurs on both subjects simultaneously
b = Number of cases in which the presence of the band occurs only in individual i
c = Number of cases in which there is only the presence of the band in the individual j
A simplified representation of the similarities was obtained by constructing dendrogram by UPGMA method (Unweighted Pair-Group Method with Arithmetic mean).13 The analysis of similarity and clustering were performed with software using the Software NTSYS pc2.1 program (Numerical taxonomy and multivariate analysis system in PC).14 By the method of Mojena,15 the dendrogram was cut at the point of θ = 0.44 (general criteria: θ-k = α +1.25σ-α). Where, θ = correction for trend lag in stages, α = value of criterion in stages (unbiased standard deviation) and K= mean of α distribution.
Genetic diversity among the genotypes
Measurements of genetic diversity in crops have important implications for plant breeding programs and the conservation of genetic resources. In the present study, genetic variation among C. canephora clones was analyzed.
The relationships among C. canephora accessions were evaluated with UPGMA clustering technique (Figure 1). The greatest similarity was between 34 and 31 (98%). The cophenetic correlation coefficient was r = 0.98, indicating a good fit. By the method of Mojena.15 it is observed that it is possible to cut in the dendrogram at the point of θ = 0.44 (general criteria: θ-k = α +1.25σ-α), indicating that the ideal number of groups should be equal to six. Hence, the dendrogram divided the genotypes into six groups, with group I composed of twenty one clones; group II composed by only one; and group III composed only one, the fourth is composed by one clone; the fifth consists of by twenty four; and the sixth by two clones (Table 2). Based on the similarity matrix and dendrogram, we can infer that there is great genetic diversity among clones of C. canephora and that these genetic materials are promising for use in breeding programs. However, not all clones from the same varietal group were clustered in just one group.
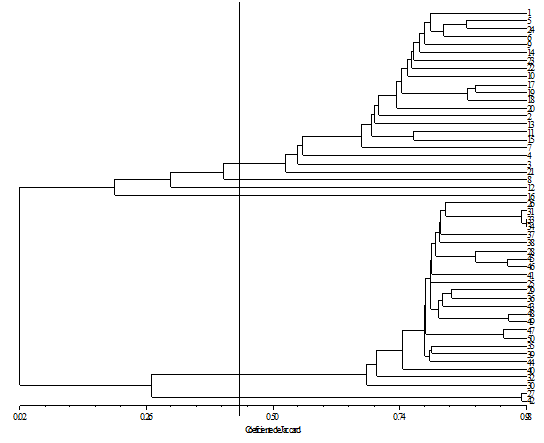
Figure 1Dendrogram of genetic similarity among 50 cultivars C. canephora, obtained from SNPs markers, using the UPGMA method.
Groups |
Genotypes |
1 |
1, 5, 24, 6, 9, 14, 23, 22, 10, 17, 19, 18, 20, 2, 13, 11, 15, 7, 4, 3 and 21 |
2 |
8 |
3 |
12 |
4 |
16 |
5 |
26, 31, 33, 34, 37, 38, 28, 45, 46, 41, 25, 29, 36, 43, 48, 49, 47, 50, 35, 39, 44, 40, 32 and 30 |
6 |
27 and 42 |
Table 2 Grouping Coffea canephora clones using Unweighted Pair-Group Arithmetic Mean Method (UPGMA). 1 to 24 = Conilon and 25 to 50 = Robusta
According to the result, genotypes which showed the most similarity pair were 34 with 31 and 33, 31 with 33, 49 with 48,46 with 45 (Table 3). Moreover, the phenotypic data from field evaluation showed that low variability among clones. On other hand, SNPs markers were able to classify the two varieties clearly implying that Robusta and Conilon are divergent heterotic groups with complementary characteristics. Thus, this marker revealed a high degree of polymorphism, which allowed a satisfactory understanding of genetic diversity among the C. canephora accessions. Moreover, they allowed the proper identification of the main current phenotypic varietal groups and showed to be able to resolve doubts about the accession classification. This ability represents a great advantage, because the high intra-specific variability and the environmental effects produce a great amount of types, which can hinder the identification of Robusta and Conilon based only on the phenotypic evaluation.
Clones |
Clones |
Jaccard similarity coefficient |
34 |
31 |
0.9889 |
34 |
33 |
0.9868 |
31 |
33 |
0.9854 |
49 |
48 |
0.9651 |
46 |
45 |
0.9618 |
46 |
28 |
0.9167 |
24 |
5 |
0.9157 |
19 |
17 |
0.9135 |
29 |
28 |
0.9125 |
18 |
17 |
0.9107 |
18 |
19 |
0.89917 |
36 |
29 |
0.8816 |
30 |
26 |
0.8813 |
Table 3 Genetic similarity among 13 clones of C. canephora based on SNPs molecular markers calculated using the Jaccard coefficient.
The present study adds a SNP markers system to the marker repertoire of C. canephora, which is cost-effective and highly flexible. It clearly revealed that there are dissimilarities between of Conilon and Robusta. Within Robusta variety, clones presented a more similarity that is, almost grouped in group 5. However, Prakash et al..4 reported a high amount of diversity in a sample of Indian Robusta coffee. However, the Conilon showed more divergence. Thus, increasing the use of Conilon accession in breeding program would have a lot of benefits. Moreover, the exploitation of heterosis resulting from crosses of the two groups can be very advantageous. This strategy has been used successfully in some breeding programs around the world.16,17
Furthermore, Conilon and Robusta have complementary characteristics. Robusta plants present high resistance to rust and nematodes, and give good beverage. Whereas, Conilon plants are tolerant to drought and they are easier to cultivate due to smaller size. So, these populations compose a perfect combination to use in a reciprocal recurrent selection program, as it has been already performed in Ivory Coast, since 1984.17-19 Although higher genetic gains could be obtained using Guinean versus Congolese strategy, considerably progress has been also achieved between Congolese subgroups.18
Even though, clones used in this study represent a minimal part of the natural variability of C. canephora germplasm, SNPs markers were able to classify the two clones clearly implying that Robusta and Conilon are divergent groups with complementary characteristics.
My special thanks go to Third World Academy of Sciences (TWAS)-Italy and Conselho Nacional de Desenvolvimento Cientifico e Technologico (CNPq)-Brazil, for the financial support.
There is no conflict of interest.
None.

©2017 Bikila, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.