Journal of
eISSN: 2373-437X


Research Article Volume 11 Issue 1
1Facultad de Ciencias, Universidad Autónoma de Baja California (UABC)
2Laboratorio de Genética, Fisiología y Reproducción, Facultad de Ciencias, Universidad Nacional del Santa, Nuevo Chimbote, Ancash, Perú
Correspondence: Amelia Portillo López, Facultad de ciencias, Universidad Autónoma de Baja California. Km 103 carretera Tijuana-Ensenada. Ensenada, B.C, CP 22860, México, Tel 526461528211
Received: January 26, 2023 | Published: February 6, 2023
Citation: González-Martínez S, Portillo-López A, López-Landavery EA, et al. Response of the obligate halophile fungus Aspergillus loretoensis to stress salinity. J Microbiol Exp. 2023;11(1):26-33. DOI: 10.15406/jmen.2023.11.00382
Fungi are recognized as indigenous microbes in natural hypersaline habitats. Aspergillus sp, among other fungi, is predominant in those environments; however, their adaptative abilities are recently studied. This study analyzes the transcriptomic response of an obligate halophile Aspergillus loretoensis under two salinity conditions (4% and 15% NaCl). This fungus shows stress under the low NaCl concentration tested since it overexpresses genes like SOD2 (oxidative stress and oxygen toxicity), ASG (resistance to salinity), and transmembrane transport (ZRT2, OAC1, PMA1, ZRC1, SNQ2, MCH4, YO075, SIT1). Meanwhile, at 15% NaCl, the up-regulated genes at 15% NaCl were related to osmolytes transport (STL1, HXT13, ZRT1), carbohydrate transport, and metabolism (MAL11, PK1, ITR1), all suggesting their adaptive conditions. This fungus expresses interesting metabolic enzymes with potential uses in biotechnology as invertases, isomerases, maltases, and lipases. As well it showed biosynthetic pathways related to oil degradation and antibiotic production.
Keywords: glycerolipids, marine fungi, gene expression, osmoregulation
Aspergillus loretoensis was recently described as an obligate halophile species, it was found in the deep sediment of the ocean where the salinity is around 3.5% (0.6M) even when this fungus can grow at 25% (4.31 M) of NaCl, showing an optimal of 15% (2.58 M).1 Halophiles "salt-loving" require salt to grow and they are classified as non-halophiles that require less than 0.2 M of NaCl but can tolerate higher salinities and are considered halotolerant; as the extremely halotolerant black yeast Hortaea werneckii. Then the slight halophiles that show grow at 0.2 M to 0.5 M of NaCl, characteristic of marine microorganisms. The next group is the moderate halophiles that grow at 0.5 M to 2.5 M NaCl and the extreme halophiles above 2.5 M NaCl.2 Halophile fungi have been described since 1998, and more species in nature are discovered yearly.3-5 To date, there have been found few obligate halophiles fungi among them; Aspergillus baarnensis,6 Gymnascella marismortui,3 Wallemia ichthyophaga,7 A. penicillioides, A. unguis,8 A. salinarum,9 A. ruber,10 W. muriae,7 Gymnoascus halophilus,11 A. salisburgensis, A. atacamensis,12 A. sydowii,13 A. montevidensis,14 A. loretoensis1 among others. Halophile fungi are less studied than the prokaryote part; however, they are an excellent source to understand the eukaryote mechanism to bear the cell osmolarity. These organisms are good candidates as industrial enzymes, anticancer, antimicrobial compounds, antioxidants for food prevention, and cosmetics industry producers.15,2 In the fungi kingdom, there are several adaptations to osmotic pressure; the main is "salt in," where the cell accumulates high concentrations of inorganic ions to obtain osmotic balance K+ ion is the most important and excludes the toxic Na+ ion.16 Another strategy is "organic osmolytes" (low-salt, salt-out, or compatible solute), where microorganisms use organic solutes to maintain the osmotic balance. Those solutes are polyols such as glycerol, arabitol, and mannitol, preventing the intercellular sodium below toxic levels.16,17 Other osmotic protection mechanisms include changes in the cell wall thickness, pigmentation, fatty acid accumulation, amino acids, and soluble sugar production.17
A. loretoensis is capable of growth at 4.31 M NaCl; however, the strategies to be an obligate halophile are unknown, which is why this study focuses on analyzing the transcriptomic response of A. loretoensis under two salinity conditions (4 and 15% NaCl) to find out which genes are involved in its halotolerant adaptations. This work contributes to understanding the metabolic process of A. loretoensis as a new species of California Gulf sea habitant.
Samples and total RNA extraction
A. loretoensis was isolated from marine sediments at 275 m deep in Loreto Bay, Baja California Sur, México (coordinates: 25°89'14" N 111°25'09" W). The isolation methodology was described previously.18 The isolate was deposited at the CM-CNRG collection from INIFAP (Colección de Microorganismos del Centro Nacional de Recursos Genéticos, Instituto Nacional de Investigaciones Agrícolas y Pecuarias, México (https://vun.inifap.gob.mx)), and the isotype culture to Herbario BCMEX-UABC fungi collection, with accession number 6006. The accession number at INIFAP is CM-CNRG 624 holotype culture. A. loretoensis also was registered at mycobank (http://mycobank.org/page/Simple%20names%20search) with accession number 830181, and it is associated to the Index Fungorum (http://www.indexfungorum.org/names/NamesRecord.asp?RecordID=830181).
A. loretoensis was cultured in the dark for seven days in YPD (Yeast Extract-Peptone-Dextrose) broth at 4 and 15% NaCl at 28°C, with three replicates. Then, the mycelium was collected and immediately frozen with liquid nitrogen, lyophilized, and homogenized using a pestle and mortar. Total RNA was purified using the EZNA Fungal RNA mini kit (OMEGA®). RNA integrity was confirmed by electrophoresis (1% agarose gel and TAE 1X). Residual DNA was eliminated by RQ1 RNase-Free DNase I kit (Promega, Madison, WI, USA). RNA was quantified using a QubitTM RNA Broad Range assay (Life Technologies, USA), and their quality was confirmed using RNA 6000 Nano Kit (Agilent Technologies, Wilmington, DE, USA). All samples had an RNA Integrity Number (RIN) > 7 and were considered for library preparation.
Libraries construction and sequencing
TruSeq® RNASeq v2 (Illumina, San Diego, CA, USA) was used to construct the libraries. The libraries concentration was quantified with the Qubit® dsDNA Broad Range assay (Life Technologies, USA), and the cDNA fragments were verified in a 4% agarose gel. Each library's size was determined on an Agilent 2100 Bioanalyzer® using the Agilent DNA 1000 kit (Agilent Technologies, Germany). The final concentration of libraries and sequencing control (PhiX) was 10 pM and 1%, respectively. The MiSeq® System Reagent V3 kit was used for sequencing (Illumina MiSeq platform), generating pair-end reads of 76 bp.
De novo transcriptome assembly
The quality of the raw reads obtained from the MiSeq sequencer was verified by FastQC V0.11.7.19 The preprocessing of reads was carried out with the Trimmomatic v0.36.20 In this phase, adapters and sequences with ambiguous 'N' bases and quality values < 30 were removed and reads with a length less than 36 bp. Before assembly, orphan reads were removed, and all left and right pair-end reads were concatenated. De novo assembly of linked reads was performed with Trinity v2.6.6 24 with default parameters.21 The assembled transcriptome quality was assessed with Transrate v1.0.3,22 and Trinity utilities, while its completeness with BUSCO v4.0.2 based on fungi_odb10 database.23
Transcripts abundance and differential expression analysis
De novo transcriptome from Transrate and reads from Trimmomatic were used to calculate the abundance of transcripts from each replicate. All reads were aligned against the indexed transcriptome using Bowtie2 v2.3.5.24 The abundance was calculated using the RNA-Seq by expectation-maximization algorithm included in Trinity (RSEM).25 The differential expression analysis was based on the raw counts from replicates, and the edgeR package v3.28.1.26 Counts were normalized, considering the library size. Then, low abundance transcripts (CPM < 0.9) in all three replicates were removed. For statistical analysis, a binomial generalized linear model was fitted to the normalized counts for pairwise comparison between NaCl concentrations. A multiplicity correction to the control (the expected false discovery rate (FDR)) was made through the Benjamini-Hochberg method.27 Transcripts with an adjusted P-value (FDR) < 0.01 and at least a two-fold change (FC > |2|) were considered differentially expressed transcripts (DET).
Functional annotation and enrichment analysis
The functional annotation of all transcripts was made through a local BLAST with the NCBI-BLAST-2.4.027 and the Swissprot/Uniprot databases,28 based on release 2020_2. Trinotate v3.0.1 software29 was used to assign the best BLAST result for each protein against the SwissProt/UniProt database and PFAM predictions domains. Gene ontology (GO) was analyzed based on the more significant categories between differentially expressed transcripts (DET), using the R packages biomaRt v2.42.1,30 cluster Profiler v3.14.3,31 and the gene set enrichment analysis strategy (GSEA) through the correction of FDR < 0.05. The enricher function identified KEGG pathways from DETs lists from the cluster profile R package and edition of FDR < 0.1. Enrichments for GO and KEGG analysis considered all annotated transcripts as background. Finally, the predicted interactions between the proteins encoded by DET between treatments were modeled using the STRING v11.0 online tool.32 Saccharomyces cerevisiae was used as a reference to set the predicted interactions based on the mean network edges confidence (0.400) for their interaction score. Nodes without connection were removed.
Transcriptomic analysis and quality assessment
The transcriptome of A. loretoensis generated 12 832 262 paired-end raw reads for 4% NaCl treatment and 10 986 333 paired-end raw reads for 15% NaCl treatment. The read length was 35-76 bp, and GC content of 60% for both conditions. After the quality control of raw reads with Trimmomatic, the initial transcriptome assembly generated 15 123 transcripts with 10,507 putative genes. The median contig length was 1,525 bp with N50 of 2 897 bp and GC content of 56.60% from 28 887 258 total assembled bases. The longest isoform per putative gene median size was 1 175 bp with N50 of 2 475 bp. The assembly quality assessment with Transrate indicated that 11 354 transcripts and 7 985 putative genes were retained, representing 75 and 76%, respectively. Statistics based on all transcripts from clean assembly reported a median contig length of 1 734 bp, a GC content of 56.69%, and N50 of 2 831 bp. The median contig length was 1 540 bp with an N50 of 2 561 bp (Table 1). The overall alignment rate (mapping) generated with bowtie2 between the processed reads against the assembled transcriptome was 96.81 and 97.70% for 4 and 15% NaCl treatment, respectively. On the other hand, BUSCO results generated a high integrity score, with 723 complete orthologous groups from 758, representing 95.4%. Single copies represented 70.5%, and 29.5% were duplicated.
Final assembly |
|
Total Trinity 'genes' |
7,985 |
Total Trinity transcripts |
11,354 |
GC (%) |
56.69 |
Stats based on all transcript contigs |
|
Contig N50 (bp) |
2,831 |
Median contig length (bp) |
1,734 |
Average contig (bp) |
2,088 |
Total assembled bases |
23,705,699 |
Stats based on only longest isoform per 'gene' |
|
Contig N50 (bp) |
2,561 |
Median contig length (bp) |
1,540 |
Average contig (bp) |
1,878 |
Total assembled bases |
14,997,382 |
Assembly quality assessment |
|
Recovered transcripts with Transrate (%) |
75.1 |
BUSCO completeness (%) |
95.4 |
Overall alignment rate (%) |
|
4% NaCl reads |
96.81 |
15% NaCl reads |
97.7 |
Annotation |
|
Transcripts with coding region |
10,137 |
Transcripts with BLAST hit (BLASTX) |
8,162 |
Transcripts with BLAST hit (BLASTP) |
6,797 |
Transcripts with orthologous groups (ENOG and COG) |
1,474 |
Transcripts with KEGG pathway |
5,948 |
Transcripts with GO terms |
6,972 |
Transcripts belonging to protein families (pfam) |
4,449 |
Table 1 Statistical summary for the de novo transcriptome assembly and annotation from 4% and 15% NaCl libraries of Aspergillus loretoensis
Functional annotation of the whole transcriptome
All transcripts were annotated with the Trinotate pipeline. The sprot_Top_BLASTX_hit results showed an annotation level of 71.8%. (Table 1) Based on the eggnog database, 13% of transcripts were annotated. Eggnog reports annotations with two categories: ENOG and COG (Clusters of Orthologous Groups), with COG composed of twenty-five categories. The most abundant COG categories for all transcripts were carbohydrate transport and metabolism, post-translation modification, protein turnover, chaperones, amino acid transport and metabolism, and lipid transport and metabolism. Annotation from the KEGG Pathway database showed that 52.4% of transcripts had a match. Several metabolic pathways were considered essential to halophilic as glycerolipids, starch, and sucrose synthesis. Metabolic analysis shows several enzymes related to sucrose and starch metabolism where we can find invertase, maltase, and isomerase, which are important to industrial processes (Table 2). Also, genes were found in antibiosis biosynthesis pathways such as streptomycin, penicillin, neomycin, hydrocarbon degradation, and vitamin and hormone producers (Table 3).
Glycerolipid metabolism |
Sucrose and starch metabolism |
|
Enzyme |
name |
Código de la enzima Nombre |
ec:2.7.1.107 |
kinase (ATP) |
ec:2.7.1.2 glucokinase (phosphorylating) |
ec:3.1.3.81 |
diphosphate phosphatase |
ec:2.7.1.1 hexokinase type IV glucokinase |
ec:2.7.1.29 |
kinase |
ec:2.7.1.4 fructokinase (phosphorylating) |
ec:2.3.1.158 |
acyltransferase |
ec:3.1.3.12 trehalose-6- phosphatase |
ec:3.1.3.21 |
alpha-glycerophosphatase |
ec:2.7.7.9 uridylyltransferase |
ec:3.1.1.23 |
lipase |
ec:3.2.1.4 endo-1-4-beta-D-glucanase |
ec:1.1.1.156 |
2-dehydrogenase (NADP+) |
ec:3.2.1.3 1-4-alpha-glucosidase |
ec:3.1.1.3 |
lipase |
ec:3.2.1.1 glycogenase |
ec:2.3.1.51 |
O-acyltransferase |
ec:5.3.1.9 isomerase |
ec:3.1.3.4 |
phosphatase |
ec:3.2.1.48 alpha-glucosidase |
ec:1.1.1.2 |
dehydrogenase (NADP+) |
ec:3.2.1.91 1-4-beta-cellobiosidase (non-reducing end) |
ec:1.2.1.3 |
dehydrogenase (NAD+) |
ec:5.4.2.2 (alpha-D-glucose-1-6-bisphosphate dependent) |
ec:3.2.1.22 |
melibiase |
ec:3.2.1.20 maltase |
ec:1.1.1.21 |
reductase |
ec:3.2.1.21 gentiobiase |
ec:2.4.1.34 synthase |
||
ec:3.2.1.28 trehalase |
||
ec:3.2.1.26 invertase |
||
ec:2.4.1.1 phosphorylase |
||
ec:2.4.1.18 branching enzyme |
||
ec:2.4.1.15 synthase (UDP-forming) |
||
ec:2.4.1.11 synthase |
||
ec:2.4.1.25 disproportionating enzyme |
||
ec:2.4.1.21 synthase (glycosyl-transferring) |
||
Table 2 Enzymes involved in glycerolipid, sugar and starch metabolism
KEGG Pathway |
Pathway ID |
Genes found in A. loretoensis transcriptome |
Antibiotics |
|
|
Biosynthesis of antibiotics |
map01130 |
129 |
Biosynthesis of streptomycin |
map00521 |
5 |
Biosynthesis of novobiocin |
map00401 |
3 |
Biosynthesis of neomycin, kanamycin and gentamicin |
map00524 |
2 |
Biosynthesis of monolactamics (Tabtoxinine β-lactam) |
map00261 |
2 |
Biosynthesis of carbapenem (Nortienamycin) |
map00332 |
2 |
Biosynthesis of penicillin and cephalosporin |
map00311 |
1 |
Hydrocarbons degradation |
||
Degradation of toluene |
map00623 |
3 |
Degradation of xylene |
map00622 |
1 |
Degradation of chlorocyclohexane |
map00361 |
1 |
and chlorobenzene |
||
Vitamins |
||
Metabolism of riboflavin |
map00740 |
7 |
Metabolism of B6 vitamin |
map00750 |
4 |
Metabolism of xenobiotics |
||
Metabolism of xenobiotics, cytochrome P450 |
map00980 |
4 |
Hormone biosynthesis |
||
Biosynthesis of steroids |
map00100 |
4 |
Biosynthesis of steroid hormone |
map00140 |
3 |
Pharmaceutic metabolism |
||
Metabolism of fluorouracil (chemotherapeutic) |
map00983 |
5 |
Metabolism of azathioprine 6-Mercaptopurinan |
map00983 |
4 |
Metabolism of isoniazid (tuberculosis treatment) |
map00983 |
3 |
Metabolism of irinotecan (cancer treatment) |
map00983 |
1 |
Other compounds degradation |
||
Degradation of benzoate |
map00362 |
7 |
Degradation of naphthalene |
map00626 |
1 |
Table 3 KEGG pathways of biotechnological interest were found in the transcriptome of Aspergillus loretoensis under two NaCl treatments (4 and 15%)
Sixty-one percent of transcripts were annotated in the gene ontology database (GO), generating 48,175 hits. In the biological process category, the most abundant genes were transmembrane protein and pathogenesis. ATP and metal ion binding (zinc) were the most abundant for the molecular function. The tops of the cellular component category were the nucleus and cytosol. Transcripts from A. loretoensis had homology mainly with species of Schizosaccharomyces sp., Aspergillus sp., Saccharomyces sp., and Neurospora sp.
Differential expression analysis and gene enrichment
Early-stage differential expression analysis removed 1,400 transcripts with low expression through all replicates (CPM < 0.9). A comparison of replicates consistency evaluated with multidimensional scaling showed that each NaCl treatment clustered separately (Figure 1a). The standard dispersion value, an indicator of the biological coefficient of variation (BCV), was 0.34. Mathematically, the BCV is equal to the square root of typical dispersion and had a value of 0.58 (Figure 1b). Statistical comparison between 15% NaCl against 4% NaCl generated 420 DET with FDR < 0.01 and two-fold change (FC > |2|) parameters. Regarding 15% NaCl treatment, two hundred and twenty-seven transcripts were up-regulated, while hundred and ninety-three transcripts were down-regulated. DET distribution between NaCl treatments was represented on mean average (MA) and Volcano plots (Figure 1 c-d). The heatmap, constructed with DET, displayed the aggrupation of replicates within its corresponding cluster and confirmed different expression patterns between each NaCl treatment (Figure 2).
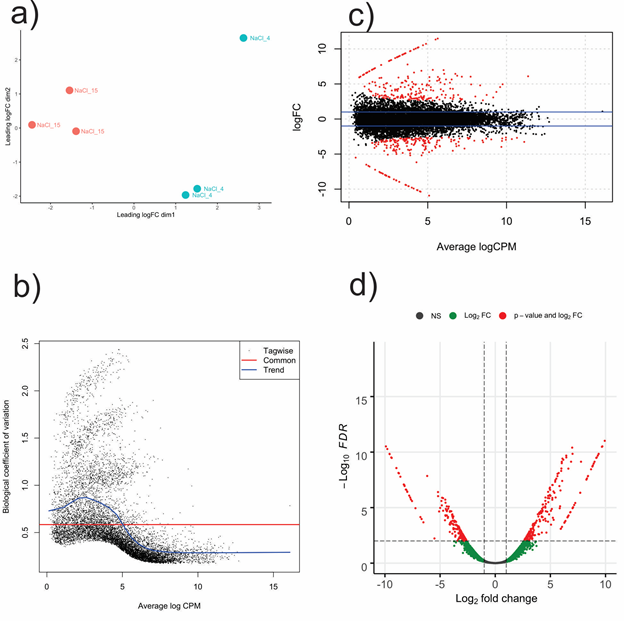
Figure 1 Differential expression analysis with edgeR in Aspergillus loretoensis. 4% NaCl treatment was used as control during the comparison. A) MDS plot based on the TMM (Trimmed Mean of M-values) normalized RNA libraries. B) Scatterplot for the biological coefficient variation between NaCl treatments. C-D) MA plot (upper) and Volcano plot (lower) showing the spatial distribution of differentially expressed transcripts. 15: 15%, 4: 4%, NS, non significant; FC, fold change; FDR, false discovery rate.
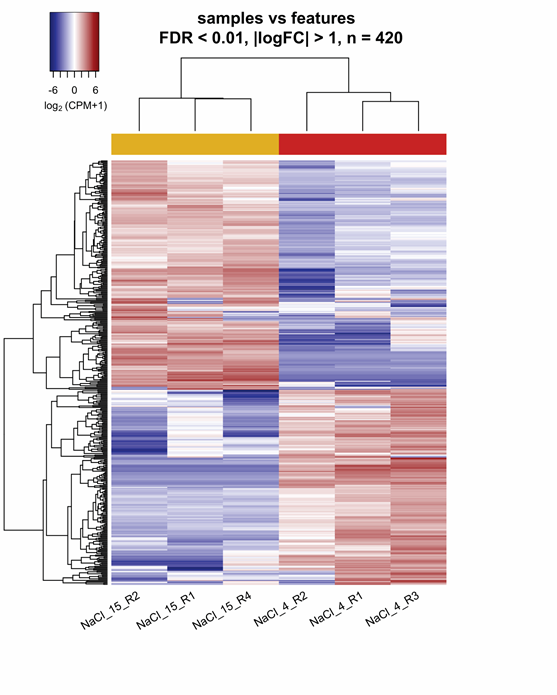
Figure 2 Clustering of the up and down-regulated transcripts from 15% NaCl vs 4% NaCl comparison in Aspergillus loretoensis. FDR, false discovery rate; FC, fold change; CPM, counts per million; Red: Up-regulated, Blue: Down-regulated, R, replicate.
A gene enrichment analysis was done based on DET to know the overrepresented biological categories and KEGG pathways. GO enrichment profile for 4% NaCl included terms related to the mitochondrion, oxidation-reduction process, ribosome, cytoplasmic translation, protein kinase activity, and transmembrane transporter activity (Figure 3). Compared with 4% NaCl, the GO enrichment profile for 15% NaCl was broader. It included terms related to nuclear chromatin, cellular response to DNA damage stimulus, mRNA processing, protein ubiquitination, RNA splicing, transferase activity-transferring glycosyl groups, carbohydrate metabolic process, and hydrolase activity-acting on glycosyl bonds. One aspect to highlight is that transcriptional activity was highly enriched at 15% NaCl treatment, as evidenced by the terms associated with RNA Polymerase II and methylation, probably as a response to high salinity. Regarding KEGG enrichment, the profiles included different pathways in a general way, indicating a specific response to NaCl treatments. Enriched KEGG pathways at 4% NaCl were related to nicotinate and nicotinamide metabolism, necroptosis, citrate cycle (TCA cycle), SNARE interactions in vesicular transport, and PPAR signaling pathways, among others. At 15% NaCl, the paths were related to the metabolism of sugars, amino acids, and cell size control. The most significant and abundant pathways at 15% NaCl included glucagon, hippo, neurotrophin, amino sugar, nucleotide sugar, arginine, and proline metabolism. The protein-protein network constructed in the STRING online database and based on S. cerevisiae confirmed the enrichment analysis results. At 4% NaCl, proteins with high confidence were related to the oxidation-reduction process and stress (HSP78, HSP30, FES1, PMA, RG11, YOR1, HYR1, MDR1), TCA cycle (SDH1 and UGA2), and carbohydrate metabolism and transport (GAL, STL1, GLC3). Proteins at 15% NaCl were related to transcription (STP15, MOT2, MED7, FCP, NRD1, ESS1), methylation and RNA processing (RSC8, NOP1, UTP6, MES1, LSM5), cell division (NUF2, MAD2, APC2, MOB1), electron transport chain (COX1, COX2, CYB2) and metabolism and transport of carbohydrates (PK1, ITR1).
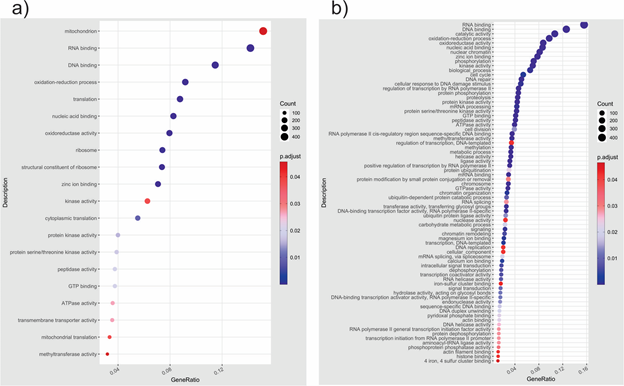
Figure 3 GO enrichment based on differentially expressed transcripts from a) 15% NaCl vs b) 4% NaCl comparison in Aspergillus loretoensis.
Data availability
Data supporting this work is available at GenBank. All RNA-Seq raw reads were deposited into the Sequencing Read Archive (SRA) of NCBI with the accession numbers from SRR15465818 to SRR15465823. The BioProject ID of our data is PRJNA739890, and the BioSample accession is SAMN19773222. The Transcriptome Shotgun Assembly (TSA) project has been deposited at DDBJ/EMBL/GenBank under the accession GJLJ00000000. The version described in this paper is the first version, GJLJ01000000.
Previously it was reported that the morphological changes that show A. loretoensis when it was exposed to 4%, 15%, and 25% of salinity, where the mycelia were scarce at 25% NaCl. It shows circles in the hyphae morphology; however, 15% was the optimal growth; those observations were taken in a count to choose 4 and 15% to analyze the transcriptome.1
The putative genes found in A. loretoensis (7 985 to 10 507 genes) are within the range of the genes reported for other Aspergillus sp. as A. fumigatus (10,087 genes), A. nidulans (10 937 genes), A. niger (14 504 genes), A. oryzae (12,322 genes), A. montevidensis (12 296 genes), A. sydowii (11 269 genes). Genome and transcriptome of some subgenus Polypaecilum have been reported, as A. sclerotialis (11 307 genes) with the highest gene numbers and A. salisburgensis with the lowest (8 895). The most halophilic fungus reported Walemia ichthyophaga shows a reduction in its genome with 538 genes, contrary to all fungi mentioned before.33-34 This condition is unrelated to the halophile characteristics in Polipaecilum subgenus. A. loretoensis was isolated from the sediment of the ocean. We expect that 4% salinity will be the best for this fungus however been demonstrated that 15% is the optimum for growth. The transcriptome analyses corroborate these findings with apparent differences in the gene expression related to their strategies to adapt and survive in the seawater. In the GO, it is observed that more transcripts are related to integral components of the membrane; other studies have reported the essential role that plays the features of the membrane in the osmolarity regulation as an adaptive strategy in the halophilic organism.33-35 Up-regulation of the carbohydrate metabolic process at 15% of salinity includes the glycolysis pathway, where glycerol is produced between their primary metabolites and is a compatible solute to survive extreme conditions. Interestingly, at 15% NaCl, the cell responded to DNA damage stimulus, and DNA repair is increased and not to 4%, where the fungus is stressed. Besides the physiological indication, the differential analysis indicates that A. loretoensis is stressed at 4% NaCl since several heat shock proteins (HSP) are up-regulated at this condition as HSP78, HSP30, and HSP70. Those proteins were reported to be related to osmotic stress in other fungi.36-38 OpS6 is a stress-related protein up-regulated in this condition.
This protein is a Glutathione S-transferase-like protein that detoxifies reactive electrophilic compounds; this protein protects the cells against oxidative stress by scavenging any leaked free radical present by activating the thiol group glutathione.39 In contrast, no HSP was found in the 15% NaCl condition. Transcripts of cell wall remodeling protein as Class III chitinase (CHI33) were observed in other halophilic fungi,33-35 and it was overexpressed in A. loretoensis in 15% NaCl condition. Chitinases from this family were found in Trichoderma sp. under stress conditions (low or high temperatures and carbon or nitrogen starvation).40-41 Several proteins involved in chitin and chitosan degradation were up-regulated, including endochitinase 1, endochitinase B, endochitinase B1, endo-chitosanase, and chitotriosidase-1. The up-regulation of these proteins was observed in the halophile fungi A. sydowii.35 However, a putative glycosidase YHZ7 is involved in the cell wall polysaccharide metabolic process. It seems that overexpression of related proteins protects the cell against osmotic stress. For example, S. cerevisiae mutants of CRH glycosidase genes reduced their survival in a high salt concentration.42 On the other hand, A. sydowii overexpresses the crh glycosidase gene at 2.0 M NaCl (11.6 % NaCl).35 In contrast, A. loretoensis at 4% NaCl overexpress proteins responsible for synthesizing new cell walls like ecm33, chitin synthase 8, and chitin synthase G, compared to A. sydowii where just one gene was found strongly downregulated in the presence of salt.35
The transporter systems in halophilic fungi regulate the cation homeostasis of alkali cation levels and eliminate toxic ion levels such as sodium.35 A. loretoensis overexpressed at 15% NaCl several transporters as MFS (Major facilitator superfamily): MFS10, MFSA, and ABC multidrug transporter: MDR1 and MDR2. In contrast, at 4% NaCl A. loretoensis also overexpress other MFS and ABC transporters like the MFS_sugar_transport-like MFSB, MFS-type transporter DBAD, MFS hexose transporter KHT2, uncharacterized transporter from the MFS protein family YN43, ABC multidrug transporter MDR1, ATRF, and ABCC. The MFS transporters are not specific to molecules since they can also transport ions, carbohydrates, lipids, amino acids, and peptides. It has been reported that up to 50% of the coding genome for transporters belonging to the MFS and ABC (ATP-binding cassette) family's protein has been related to drug resistance.33 MFS transporters are down-regulated on saline stress conditions in W. ichthyophaga and A. salisburgensis; however, A. loretoensis showed an up-regulation of different transporters at NaCl concentrations tested in this study. A. sclerotialis and A. sydowii showed similar conditions at high salt concentrations.37,39 Zajc et al.33 mention that this down-regulation in halophilic fungi such as W. ichthyophaga and A. salisburgensis is because of high salinity environments species belong, where competition with other species is limited. However, A. loretoensis probably has a more competitive habitat.
Another transporter up-regulated at 15% NaCl was the ZRT1 gene, which codes for a Zn transporter protein. Zinc transporters are not specific and respond to lower ion concentrations transporting other metals under stress conditions, suggesting that regulatory mechanisms act similarly under non-optimal conditions for the cell. Up-regulation of zinc transporters has been reported in A. sclerotialis under high salinity conditions.34 Contrary to the yeast Pichia kudriavzevii, where the expression of ZRT1 has been down-regulated under saline stress conditions.43 Tafer et al. mention that this regulatory mechanism under high salinity conditions is related to oxide-reduction reactions and ion exchange during high salinity.34
The high osmolarity glycerol (HOG), a map kinase pathway activated by osmotic stress, is an essential osmoregulatory mechanism in several fungi. In A. loretoensis, HOGA1 and PHSG genes regulated by stress-response elements and the HOG MAP kinase pathway were overexpressed at 15% NaCl. In other halophilic fungi, genes of this pathway have been found in Hortaea werneckii and W. ichthyophaga. In this species, the HOG genes activate at limiting salinities.33 Genes of the HOG family were found in A. sydowii, but they were not expressed differentially under saline stress conditions,35 and in A. salisburgensis a homolog of tyrosine-protein phosphatase 3 (PTP3) was found; this protein is a repressor of the HOG family.34
The STL1 gene was up-regulated at 4% NaCl, which codes for a glycerol-gate symporter protein in the plasmatic membrane. This gene has been previously reported in W. ichthyophaga, which acts during hyperosmotic shock, preventing the exit of the cell's glycerol; under these conditions, the channels of water-glycerol-porins Fps1 are closed.33 This study finds some of the general strategies of the fungi to address the osmolarity shock in A. loretoensis being the glycerol biosynthesis, the principal metabolite regulator since it was up-regulated at 15% NaCl. However, at 4% NaCl, A. loretoensis showed stress corroborating previously reported results of their optimal growth.1 We discover interesting metabolic pathways related to antibiotic biosynthesis and biodegradation of hydrocarbon molecules that could be explored in oil bioremediation in the ocean. Also, there were enzymes of biotechnological interest (invertase, isomerase, maltase, etc.) that needed further exploration since these enzymes are used in the food industry. Previously was reported the capacity of A. loretoensis to degrade lignin and starch,1 so this characteristic is important in the paper industry. More studies are necessary to analyze the conveniences of this fungus as a producer of commercial enzymes or as an oil biodegradation organism.
We thank Consejo Nacional de Ciencia y Tecnología (CONACYT) for supporting the Ph.D. scholarship to Sophia Gonzalez-Martínez. We thank Universidad Autónoma de Baja California and CICESE for allowing us to use their facilities.
The author declares that there is no conflict of interest.

©2023 González-Martínez, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.