Journal of
eISSN: 2373-437X


Review Article Volume 1 Issue 2
Chemical Biological Directorate, Defense Threat Reduction Agency, USA
Correspondence: Akbar S Khan, Senior Microbiologist and Program Manager, Chemical Biological Directorate, Defense Threat Reduction Agency, Fort Belvoir, Virginia, 22060, USA, Tel 703-767-3391, Fax 703-767-1892
Received: April 18, 2014 | Published: May 23, 2014
Citation: Khan AS. Rapid advances in nucleic acid technologies for detection and diagnostics of pathogens. J Microbiol Exp. 2014;1(2):56-61. DOI: 10.15406/jmen.2014.01.00009
In this area of detection and diagnostics, traditional culture-based methods are simple to use, relatively inexpensive but too time consuming for high-throughput testing and too complex for analysis of samples with a mixture of organisms and provide little clinical information regarding the pathogen such as antibiotic resistance genes, virulence factors or strain subtypes. Nucleic Acid based technologies such as polymerase chain reaction, DNA microarrays, and high throughput automated sequencing methods have advantages over these culture based technologies which provides more molecular information as discussed above. In the last 10years, there is rapid advancement in these nucleic acid based technologies and in this review article; I will describe the current state of the art in the nucleic acid technologies for detection and diagnostics and future trends and advances in the field.
Keywords: nucleic acid, detection, diagnostics, polymerase chain reaction, nanotechnology
Traditionally microbiologists with classical techniques have been concerned with the tedious isolation and identification of pathogenic organisms of interest from humans whenever an outbreak happened. Typically conventional methods involve isolating the microbes of interest in pure culture and performing predetermined biochemical or immunologic tests to identify and characterize it. All these classical methods are limited in sensitivity, specificity or both. In order to improve upon these test sensitivities, to achieve faster detection times and to identify hard to culture microbes, better immunoassays were developed using specific antibodies.1,2
Histologists, cytologists, and cytogeneticists have been concerned with the preparation and staining of various tissue sections for microscopic examination. The traditional stains have allowed differentiation of various cellular components. Over theyears, special stains to improve the detection of numerous cellular components and infectious microbial agents of interest have been developed. This area of laboratory practice was one of the first areas to benefit from the advances in molecular biology over the last two decade. Nucleic acid analysis such as nucleic acid hybridization, Polymerase Chain reaction, Microarray technologies and rapid high throughput automated sequencing have attracted much attention over the past 20years and is now becoming available for clinical laboratory testing on a routine basis.3‒5 The advantages of these nucleic acid based technologies in identifying organisms without special or tedious isolation, detecting nonviable organisms, and detecting genetic mutations6 or alterations in other cellular material represent the big advantage over the traditional classical technologies.3‒5 This particular review focuses on technologies for detecting nucleic acid molecules and presents some newer and more novel methods that are being developed for possible introduction into the clinical laboratory.
Nucleic acid hybridization technologies
Nucleic Acid hybridization technologies are based on simple structure of DNA which is composed of four repeating nucleotides, adenine, guanine, cytosine and thymine. DNA is coiled to form a double helix strand and composed of two strands held together by hydrogen bonds these bonds are pH and heat labile. DNA fragments from different sources with similar sequences anneal together to form double stranded DNA and this process is called hybridization as shown in Figure 1A & 1B. For detection and diagnostic purposes, particular sequences of DNA of interest can be detected with help of a probe which is a short sequence of nucleotide bases that binds to specific regions of a target sequence of nucleotides. For detection purposes the probes can be labeled either with radiolabel isotopes, or fluorescent tags so they can be detected after the hybridization to a particular target sequence of interest. DNA or RNA fragments of interested can be detected with hybridization approaches called Southern or Northern blots respectively as illustrated in Figure 1B. In past 15years, this area has been heavily commercialized and therefore a variety commercial probes for different detection targets of interest and hybridization solutions kits exists for quick application with ease. These tools, which were once used solely in the domains of researchers and anatomic pathologists, have been demonstrated to be useful for the detection of pathogens in blood culture samples as well as in other clinical specimens.7‒11
Polymerase chain reaction (PCR) technologies
The specific DNA amplification was reported first using the Polymerase Chain Reaction (PCR) in 1987. The method was developed by Mullis & Faloona.12 PCR relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region along with a DNA polymerase (after which the method is named) are key components to enable selective and repeated continuous amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. Since its invention, the field of application has expanded enormously to include clinical, veterinary, food and environmental areas. The basic PCR technique has, in recentyears, been combined with fluorescently labeled probe hybridization in the same reaction, allowing real-time monitoring of the target amplification. This development, known as real-time PCR, has been responsible for the increase in popularity of molecular assays in diagnostic laboratories. Most real-time methods are based on the principle of fluorescence resonance energy transfer (FRET). Fluorescence resonance energy transfer occurs when two fluorescently labeled molecules are in close proximity to each other and the energy from an excited donor molecule is transferred to an acceptor molecule.
Real-time PCR platforms utilize different detection formats. Fluorescent technologies employed are either nonspecific, using dyes such as SYBR Green I or SYBR Gold, which intercalate into the PCR product during the reaction, or specific, using probes to detect specific sequence amplification in the PCR. A number of different fluorescent probe chemistries have been employed in real-time PCR assays including TaqMan® (5'exonuclease) probes, HybProbes, molecular beacons and Scorpion probes. While the mechanism of fluorescent signal generation is different for each of the probe chemistries, the fluorescent signal generated by the probes or minor groove binding dyes is directly proportional to the amount of PCR product generated.13,14 Real-time PCR is quantitative with a broader dynamic range than conventional PCR.
The most commonly used chemistries utilized in real-time platforms are the hydrolysis or TaqMan probe15 and HybProbes. A TaqMan probe consists simply of an oligonucleotide probe that has a fluorescent label attached to the 5' end and a quencher at the 3' end. Once bound to the target during the PCR reaction, the probe is cleaved by DNA polymerase, which separates the fluorescent label from the quencher. When the quencher and the label are no longer in close proximity, the signal is emitted and detected in the real-time machine. Figure 2 outlines the mechanism of action of TaqMan probes. The fluorescence is directly proportional to the amount of PCR product produced. The hybridization probe or HybProbe format was developed specifically for use on the LightCycler® instrument (Roche, Basel, Switzerland). In this detection format, two separate probes are designed to hybridize next to each other on the target DNA. HybProbes are most commonly used for melting curve analysis, which is based on the probes dissociating from the target DNA at a certain temperature. Numerous protocols are described in the literature for the use of hybridization probes.16 Another chemistry less often utilized but none the less extremely specific is molecular beacons. These are probes that form a stem–loop structure when in solution and emit a signal only when bound to their specific target.17 An example of a commercial assay using molecular beacons is the methicillin-resistant Staphylococcus aureus (MRSA) assay from Cepheid (CA, USA), which is performed on the SmartCycler® instrument. Real-time PCR is most often used for detection and quantification of microorganisms but another use of real-time PCR is the ability to quantify and genotype genetic variations, such as single nucleotide polymorphisms (SNPs), using a set of specific probes for each possible SNP nucleotide within the same assay. An example of a commercially available test for SNP identification would be the Factor V Leiden test from Roche.
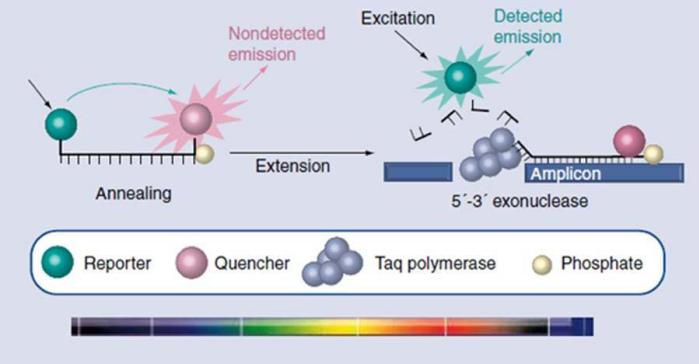
Figure 2 Schematic of TaqMan® probe technology.36
There are several advantages of real-time PCR including that it is a rapid, accurate and highly sensitive technique and can be utilized for both qualitative and quantitative analysis, and for genotyping (Table 1). These advantages have led to an explosion in the development of real-time PCR assays for diagnostic purposes driven by the need for faster and more accurate diagnostic tests. Amplification-based strategies enable the direct detection of microbial pathogens in clinical samples by molecular methods (Table 2).
Amplification Technology |
Features |
Company |
PCR |
DNA/RNA, efficient, specific |
Roche Cepheid |
Nucleic acid sequence-based amplification |
RNA/DNA, isothermal, efficien |
bioMerieux |
Strand displacement amplification |
DNA, isothermal |
Becton Dickinson |
Ligase chain reaction |
DNA, specific |
Abbott |
Branched DNA amplification |
DNA/RNA, isothermal |
Chiron |
Loop-mediated amplification |
DNA, isothermal |
Eiken Chemical Company |
Rolling circle amplification technology |
DNA/RNA, isothermal |
Molecular Staging Inc. |
Transcription-mediated amplification |
DNA/RNA, isothermal, rapid |
GenProbe |
TwistAmpTM |
DNA, rapid, isothermal |
TwistDx |
Table 1 Summary of in vitro amplification technologies36
Test |
Method |
Vendor |
CMV detection |
Hybrid capture* |
Digene ( http://www.digene.com/) |
CMV typing, screening |
Hybridization |
Digene |
CMV pp67 mRNA |
NASBA |
BioMerieux ( http://www.biomerieux.com/) |
C. trachomatis DNA Test |
Hybrid capture* |
Digene |
C. trachomatis and N. gonorrhoeae screening and/or detection |
Hybridization and amplification |
Gen-Probe ( http://www.gen-probe.com/) |
C. trachomatis detection |
LCR |
Abbott Laboratories ( http://abbott.com/) |
C. trachomatis detection |
PCR |
Roche Diagnostics ( http://www.roche.com/) |
C. trachomatis and N. gonorrhoeae detection |
SDA (BD Probetec ET) |
Becton-Dickinson ( http://www.bd.com/) |
C. trachomatis detection |
TMA |
Gen-Probe |
Culture confirmation for Mycobacteria spp. ; different fungi and bacteria* |
Hybridization |
Gen-Probe |
Gardnerella vaginalis, Trichomonas vaginalis and Candida spp. |
Hybridization |
Becton-Dickinson |
HCV detection |
PCR |
Roche Diagnostics |
HCV/HIV-1 detection (blood donor centers) |
TMA |
Gen-Probe and Bayer ( http://www.bayer.com/) |
HPV DNA Test |
Hybrid capture* |
Digene |
HPV typing/screening |
Hybridization |
Digene |
M. tuberculosis detection |
PCR |
Roche Diagnostics |
M. tuberculosis detection |
TMA |
Gen-Probe |
N. gonorrhoeae detection |
LCR |
Abbott Laboratories |
N. gonorrhoeae DNA Test |
Hybrid capture* |
Digene |
Streptococcus pyogenes (Group A streptococcus) |
Hybridization |
Gen-Probe |
Table 2 US FDA-approved tests for qualitative pathogen detectiona38
*Abbreviations: C. trachomatis: Chamydia trachomatis; M. tuberculosis: Mycobacterium tubercubsis; N. gonorrhoeae: Neisseria gonorrhoeae; NASBA: Nucleic Acid Sequence-Based Amplification
High throughput automated DNA sequencing technologies
As described by Sanger et al.,18 the dideoxynucleotide termination sequencing of DNA has undergone rapid revolution with large scale production style next generation sequencing (NGS) technologies. The next generation sequencing technologies generate its templates either by clonally amplified templates originally from single DNA molecule and single DNA molecule templates. These NGS technologies are described as these methods are classified as cyclic reversible termination (CRT), single-nucleotide addition (SNA) and real-time sequencing. Sequencing by ligation (SBL), an approach in which DNA polymerase is replaced by DNA ligase, is also described. Imaging methods coupled with these sequencing strategies range from measuring bioluminescent signals to four-color imaging of single molecular events.
Cyclic reversible termination: As the name implies, CRT uses reversible terminators in a cyclic method that comprises nucleotide incorporation, fluorescence imaging and cleavage.19 In the first step, a DNA polymerase, bound to the primed template, adds or incorporates just one fluorescently modified nucleotide (box 1), which represents the complement of the template base. The termination of DNA synthesis after the addition of a single nucleotide is an important feature of CRT. Following incorporation, the remaining unincorporated nucleotides are washed away. Imaging is then performed to determine the identity of the incorporated nucleotide. This is followed by a cleavage step, which removes the terminating/inhibiting group and the fluorescent dye. Additional washing is performed before starting the next incorporation step. Figure 3B depicts a four-colour CRT cycle used by Illumina/Solexa, and Figure 3C illustrates a one-colour CRT cycle used by Helicos BioSciences (Figure 3D). The key to the CRT method is the reversible terminator, of which there are two types: 3'blocked and 3'unblocked. The use of a dideoxynucleotide, which acts as a chain terminator in Sanger sequencing, provided the basis for the initial development of reversible blocking groups attached to the 3'end of nucleotides.20,21 Blocking groups, such as 3'-O-allyl-2'-deoxyribonucleoside triphosphates (dNTPs) and 3'-O-azidomethyl-dNTPs22, have been successfully used in CRT. 3'-blocked terminators require the cleavage of two chemical bonds to remove the fluorophore from the nucleobase and restore the 3'-OH group.
Currently, the Illumina/Solexa Genome Analyzer (GA)22 dominates the NGS market. It uses the clonally amplified template method illustrated in Figure 1B, coupled with the four-colour CRT method illustrated in Figure 3B.
Sequencing by ligation: SBL is another cyclic method that differs from CRT in its use of DNA ligase23 and either one-base-encoded probes or two-base-encoded probes. In its simplest form, a fluorescently labeled probe hybridizes to its complementary sequence adjacent to the primed template. DNA ligase is then added to join the dye-labelled probe to the primer. Non-ligated probes are washed away, followed by fluorescence imaging to determine the identity of the ligated probe.24 The cycle can be repeated either by using cleavable probes to remove the fluorescent dye and regenerate a 5'-PO4 group for subsequent ligation cycles (Figure 3C) or by removing and hybridizing a new primer to the template (not shown in the figure). Life/APG has commercialized their SBL platform called support oligonucleotide ligation detection (SOLiD).5 The method uses two-base-encoded probes, which has the primary advantage of improved accuracy in color calling and SNv calling, the latter of which requires an adjacent valid color change.
Single- nucleotide addition (pyrosequencing): Pyrosequencing is a non-electrophoretic, bioluminescence method that measures the release of inorganic pyrophosphate by proportionally converting it into visible light using a series of enzymatic reactions4,25 (Figure 3A). Unlike other sequencing approaches that use modified nucleotides to terminate DNA synthesis, the pyrosequencing method manipulates DNA polymerase by the single addition of a dNTP in limiting amounts. Upon incorporation of the complementary dNTP, DNA polymerase extends the primer and pauses. DNA synthesis is reinitiated following the addition of the next complementary dNTP in the dispensing cycle. The order and intensity of the light peaks are recorded as flowgrams, which reveal the underlying DNA sequence (Figure 3A).This chemistry is commercialized by Roche/454 sequencer.
Real-time sequencing: The next technology method to hit the commercial sector is likely to be real-time sequencing, and Pacific Biosciences is currently leading this effort.26 Unlike reversible terminators, real-time nucleotides do not halt the process of DNA synthesis. Simply put, the method of real-time sequencing involves imaging the continuous incorporation of dye- labelled nucleotides during DNA synthesis.27 With the Pacific Biosciences platform, single DNA polymerase molecules are attached to the bottom surface of individual zero-mode waveguide detectors (Zmw detectors) (Figure 3E) that can obtain sequence information while phospho-linked nucleotides (box 1) are being incorporated into the growing primer strand (Figure 3E).28
The ability to rapidly and reliably obtain the nucleotide sequence of pathogenic microorganisms present in a sample with these high throughput massively parallel sequencers changed nucleic acid-based diagnostics and detection towards rapid diagnostics and detection.
Nanotechnology based technologies
Use of nanopores as biosensors, nanopores relate to a specific class of electrochemical sensor in which an electrically insulating material is permeated by one or more pores with diameters ranging from 10 to 150nM. The insulating layer may be an organic or synthetic material. For example, nanopores have been introduced into lipid bilayers using self-assembling protein tetramers.29 Other, highly innovative, work has exploited the fact that cell membranes are naturally permeated by stable nanoscale ion channels. A feature of some of these naturally occurring structures is that they have an extremely narrow diameter (such that single-stranded nucleic acid molecules can only pass through one nucleotide at a time).30,31 Finally, so called ‘solid-state’ nanopores containing membranes can be fabricated in synthetic materials such as silicon nitride. The fabrication methodologies owe much to the technologies used in microprocessor fabrication and include such techniques as ion-beam sculpting32 or electron beams.33 The nanopore- containing membrane is placed in an electrolyte solution in an electrical cell and a voltage is applied across the membrane. Detection is based upon monitoring of the ionic current through the pores. The sensing potential of these devices depends on the interruption of this ion flow due to ‘blocking’ of the opening by different biological molecules. For nucleic acid based nanopore detection, specific capture oligonucleotides are immobilized on the surface of the nanopores and used to detect complementary single stranded sequences as they pass through the opening. Electric current is used to electrophoretically move the charged nucleic acid molecules through the pores. The observation that different strands of DNA or RNA can have a different effect on the ionic current has also led directly to the field of nanopore sequencing which will be another future next generation of sequencing machine.34
With these rapid advances in nucleic acid technologies especially next generation of sequencing platforms, these can easily adapted and applied towards detection and diagnostics of pathogens. The production of large numbers of low cost reads from these sequencing platforms makes these next generation sequencing technologies to be applied for quick de novo assemblies of out breaking pathogenic bacterial and viral genomes and a quick analysis to determine its identity as done for human influenza A viruses and virulent Escherichia coli O157 strains.34,35 Moving forward these automated sequencing technologies will play a crucial role in detection and diagnostics of pathogens of future outbreaks.
None.
Author declares that there is no conflict of interest.

©2014 Khan. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.