Journal of
eISSN: 2373-437X


Research Article Volume 11 Issue 1
1Hinda and Arthur Marcus Institute for Aging, Hebrew SeniorLife, Department of Medicine, Beth Israel Deaconess Medical Center and Harvard Medical School, USA
2Solarea Bio, Inc., USA
Correspondence: Shivani Sahni, Hinda and Arthur Marcus Institute for Aging, Hebrew SeniorLife, Department of Medicine, Beth Israel Deaconess Medical Center and Harvard Medical School, USA, Tel 617-971-5382
Received: January 17, 2023 | Published: January 25, 2023
Citation: Sahni S, Schott EM, Carroll D, et al. Randomized clinical trial to test the safety and tolerability of SBD111, an optimized synbiotic medical food combination designed for the dietary management of the metabolic processes underlying osteopenia and osteoporosis. J Microbiol Exp. 2023;11(1):1-11. DOI: 10.15406/jmen.2023.11.00379
To determine the effect of a twice daily administration of SBD111 on safety and tolerability in healthy adults in a randomized, placebo-controlled trial over 28-days. Participants were randomized to either SBD111 (n=15) or placebo (n=17). The outcomes were the number, frequency, and severity of Gastrointestinal (GI) symptoms and the number and severity of adverse events (AEs) over 28-days. Stool samples were collected and analyzed at baseline, after 28- and 56-days. Groups were compared (P< 0.05) using an intention-to-treat approach. The two groups were similar at baseline. After 28-days, the presence of GI symptoms tended to be higher with SBD111 use vs placebo (P=0.08) but the total number, frequency/severity of GI symptoms did not significantly differ. The number of AEs possibly related to the study were higher with SBD111 use vs placebo (P=0.05), there were no significant differences in the mean number/severity of AEs. The majority of reported AEs were mild, some were moderate, but none were severe. There were no significant differences in alpha diversity indices between the two groups at baseline or follow-up. SBD111 strains were identified in stool, enriched metabolic pathways for menaquinone (vitamin K2) production at 28-days, and were not detected at 56-days. The relatively low frequency and mild severity of GI symptoms and AEs suggests that SBD111 at the level tested is safe for human consumption.
Keywords: medical food supplement, prebiotic, probiotic, bone, gut microbiome, older adults
Historically, probiotics have been isolated from human stool and are derived from microbes commonly identified along the gastrointestinal (GI) tract. These probiotic microorganisms offer a benefit to the host and have been studied for their positive impact on human diseases including type 2 diabetes,1 ulcerative colitis,2 neurodegenerative diseases,3 osteoarthritis,4 and others. Recently, the gut microbiome has been implicated as a potential target to improve health outcomes in osteopenia and osteoporosis.5,6 Approximately 10 million Americans over the age of 50 years are currently living with osteoporosis culminating in 1.5 million fractures annually.7,8 The current standards of care including anti-resorptive and anabolic therapies are reserved for those with high fracture risk. Very few specific interventions are available to slow age-related bone loss that ultimately leads to the need for pharmacologic therapy. Thus there is a current unmet need for safe and effective ways to favorably alter bone turnover to slow bone loss.8,9 Probiotics offer a new opportunity for the dietary management of the processes in bone that lead to bone loss, potentially lowering the likelihood that prescription medications will become necessary.
As an alternative to the traditional source of probiotics from the intestine, Solarea Bio isolated bacteria, yeast, and filamentous fungi from fruits and vegetables typical of a healthy, plant-based diet. Fruits and vegetables harbor an immensely diverse microbiome essential for the health and survival of the plant.10-12 This untapped source of microbial diversity has facilitated the development of a novel, safe, Defined Microbial Assemblage™ (DMATM) medical food product, SBD111, for the dietary management of the metabolic processes associated with osteopenia and osteoporosis. SBD111 is composed of four strains of microorganisms isolated from different food sources: Lactobacillus brevis SBS4254, Lactobacillus plantarum SBS4260, Leuconostoc mesenteroides SBS4255, and Pichia kudriavzevii SBS4263, as well as oligofructose and dried blueberry powder. Solarea Bio has generated multiple pre-clinical validations of product efficacy in mouse models of osteoporosis.13 While the SBD111 product has been tested in multiple preclinical studies and a toxicity study in rats, it has yet to be administered to a human volunteer. An adverse event profile of this product is a necessary first step to advance it to the next phase of clinical development.
The goal of this study was to evaluate the safety (primary outcomes) and GI tolerability (secondary outcomes) of daily administration of 6.1x1010 CFU/d of SBD111 (an optimized synbiotic medical food) over a 28-day period. In addition, this study aimed to evaluate the effect of SBD111 on the fecal microbiota composition and function, as well as the persistence of SBD111 product in the GI tract after a 28-day washout period (56-day). We hypothesized that twice-daily oral administration of SBD111 would be safe for human consumption and would not cause GI intolerability over 28 days. Further, we hypothesized that the SBD111 product would be identified in the stool of participants after 28-days of dosing and would dissipate over time, such that after a 28-day washout period, the genomic signal of SBD111 microbial strains would no longer be detected by metagenomic sequencing in the stool samples.
Study design and participants
This study was a sponsor-initiated, individual level, randomized, double-blind, place-bo-controlled, parallel-arm clinical trial conducted in Roslindale, Massachusetts. The study was sponsored by Solarea Bio, Inc., which also supplied the medical food and placebo supplements. This trial was registered in clinicaltrials.gov (NCT05206864). Full trial protocol can be accessed at clinicaltrials.gov. It was performed in concordance with the Declaration of Helsinki and received ethical approval from the Institutional Review Board (IRB-00054643) at Advarra. Each participant provided written informed consent. The first participant was enrolled (randomized) on September 24, 2021; data collection ended February 7, 2022. See Supplementary Table S1 for a schedule of study visits and assessments.
Eligible participants were a convenience sample of adult men and non-pregnant women aged 18-70 years, with a Body Mass Index (BMI) between 18.5 and 35 kg/m2, systolic blood pressure (SBP) ≤ 155 mm Hg, diastolic blood pressure (DBP) ≤ 95 mm Hg, not using probiotic or prebiotic supplements or antibiotic treatment, and with screening blood tests results within normal range. For further eligibility details see Supplementary Material S2. Participants were recruited via digital advertisements and study flyers from the local community. Interested adults underwent a pre-screening medical interview by telephone to review all disqualifying medical conditions and assess history of probiotic/prebiotic consumption. Those who qualified were invited for an in-person screening visit prior to randomization. During the in-person screening visit, blood pressure, height, and weight were recorded, and BMI was calculated before collecting blood to check white blood cell count, neutrophils, platelets, hemoglobin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and total bilirubin. A urine sample was collected and used for a dipstick pregnancy test for women who were undergoing screening.
Participants were also excluded if, in the investigator’s opinion, there was evidence of cognitive impairment or dementia which was sufficient to interfere with informed consent or adherence to the study protocol. The clinical site physician assessed study eligibility for all potential participants prior to collection of baseline data.
Test articles
The study medical food product (SBD111) is an orally delivered synbiotic capsule containing a mixture of live microbial probiotic strains (Lactobacillus brevis SBS4254, Leuconostoc mesenteroides SBS4255, Lactobacillus plantarum SBS4260, and Pichia kudriavzevii SBS4263) with natural prebiotics and inert formulation ingredients (Supplementary Table S3). Each microbial strain is manufactured independently as a dry bulk intermediate and subsequently blended together with the prebiotic fibers. The final formulated study product (SBD111) and placebo were prepared by Dietary Pros (7111 W. Stewart Ave, Wausau, WI 54401) and were packaged as a monthly supply of capsules in bottles, which were refrigerated until dispensed. Quality control testing of the study product was conducted by an independent, certified test lab. The test articles were provided to all trial participants upon randomization starting on September 24, 2021. Eligible adults were randomized to one of the two test articles: either Control (placebo) or SBD111 for a period of 28 days. SBD111 was supplied in a vegetable-based capsule at a dosage of 6.1x1010CFU/d total microbes and 500mg of total probiotics and prebiotics per capsule. Placebo was also supplied in a vegetable-based capsule containing 500mg of maltodextrin per capsule that were administered in identical bottles (Supplementary Table S4). Participants consumed two capsules per day, one with breakfast and one with dinner. A block randomization scheme with varying block sizes in a 1:1 ratio was used. The date of randomization marked the start of follow-up for each participant. All study products were identical in packaging and had the same appearance, taste, and texture. Trial participants, investigators, and all study staff involved in baseline and follow-up data collection were masked to participants’ randomization assignments. A data manager from the Marcus Institute who was not involved in the data analyses had the assignment codes and exclusive access to the key linking assignment with participants. Study personnel and investigators were masked until all data collection was completed. The analyst was also masked until the analysis was complete. Project Director enrolled study participants and assigned them to intervention groups based on the assignment codes.
Primary outcomes
The primary tolerability outcomes were the number, frequency, and severity of adverse GI symptoms over 28-day that are possibly or probably related to administration of SBD111 as assessed by the study physician (DPK), who was blinded to group assignment. GI tolerability was measured by a modified Gastrointestinal Tolerability Questionnaire on days 0-7, then weekly thereafter and reviewed at prescribed intervals i.e. at baseline, days 1-7, day 14, day 21, and day 28.14 We utilized a classification system for AEs such that both total as well as GI specific AEs were recorded.
Secondary outcomes
Secondary outcomes related to safety were the number and severity of all adverse events (AEs) that were possibly or probably related to administration of SBD111. Self-report of Adverse Events (AE) was performed daily and reviewed at prescribed intervals to monitor intervention side effects and was reviewed by the clinical physician. All participants were interviewed by phone using standardized symptom checklists once they self-reported an AE. Participants were asked to report any serious events within 24 hours of occurrence to the study team. The study team assessed the severity of adverse events and whether they were expected, and related to the study.
Exploratory outcomes
Exploratory outcomes included fecal microbiota composition, function, and persistence of SBD111 product in the GI tract after a 28-day washout period (56-day).
Stool collection, processing and analysis: Gut microbiome composition was monitored at baseline, after 28 days, and after 56 days by shotgun metagenomic analyses of stool samples. Participants were instructed to collect an at-home stool sample at these timepoints and to ship the samples to Solarea Bio using a prepaid shipping label. Stool collection kits were purchased from Microba Life Sciences and consisted of a swab for sample collection. All materials were pre-labeled with an assigned de-identified study ID and time interval of the sample collection, and were returned via pre-paid USPS Priority to Solarea Bio (cost-effectively at ambient temperature). Staff ensured sample intake and tracking, quality control, protection of confidentiality, aliquoting and temporary storage at -80°C. Once all stool samples were collected, Solarea Bio shipped them in bulk on dry ice to Microba Life Sciences in Brisbane Australia. Once there, Microba performed DNA extractions, library preparations, and sequencing. Raw sequencing reads were transferred to Solarea Bio for bioinformatic analyses.
DNA/RNA extraction, library preparation, and sequencing: DNA was extracted from samples using the DNeasy 96 PowerSoil Pro QIAcube HT Kit (Qiagen 47021) according to manufacturer’s instructions with a modification in the initial processing step on the QIAcube HT DNA extraction system (Qiagen 9001793). Mechanical lysis was performed with the PowerBead Pro beads (Qiagen 19311). Extracted DNA was quantitated using a high sensitivity dsDNA fluorometric assay (QuantIT, ThermoFisher, Q33120).
DNA libraries were generated using the Illumina DNA Prep (M) Tagmentation Kit (Il-lumina, 20018705) with IDT for Illumina DNA UD Index Sets A-D (Illumina 20027213-16) following manufacturer’s instructions, with a modification of volume for processing in a 384-plate format. Library quality was assessed by measuring DNA concentration using the high sensitivity dsDNA fluorometric assay (QuantIT, ThermoFisher, Q33120) and gel analysis. Individual libraries were pooled in equimolar amounts to create a sequencing pool. The pooled library was sequenced on the Illumina NovaSeq6000 using v1.5 300 bp PE sequencing reagents. DNA libraries were sequenced to a target 5Gbp per sample with a minimum of 2Gbp. Sequencing quality was assessed by Illumina sequencing metrics.
Metagenomic sequence analysis: Quality control of raw reads was performed using SolexaQA++ package14 discarding reads less than 50bps and with a Phred quality score<20. Removal of host sequencing reads was performed using Bowtie2 v 2.4.2,15 with default parameters with the reference human genome GRCh38. Nonpareil v3.016 with default parameters was used to estimate the average coverage for each sequenced library using a cutoff of 80% community coverage. One metagenomic sample showed less than 80% coverage and was excluded from further analyses. Taxonomic classification of metagenomic samples was per-formed using MetaPhlan3 with default parameters. Taxon relative abundances were calculated based on marker genes in a specific clade at different classification levels (species, genus, family). ITS coverage of P. kudriavzevii in the metagenomes was calculated by mapping the metagenomics reads to the ITS sequence (coverage (X) = number of mapped reads * read length/ ITS length). Functional profile was characterized using HUMAnN3 with default parameters and reference pathways databases including UniRef, KEGG, UniPathway, and MetaCyc. Difference in abundance in metabolic pathways was assessed across study groups and time points.
Whole genome sequencing of SBD111 microbial strains: DNA was extracted from individual colonies using the Zymo quick DNA bacterial/fungal kit (Zymo). DNA libraries were built using the Nextera XT DNA library preparation kit (Illumina Inc) following manufacturer’s instructions. Concentration of the DNA libraries was calculated using Qubit 3.0 Fluorometer (ThermoFisher scientific). Sequencing of DNA libraries was performed in an Illumina MiSeq (MiSeq Control Software v2.6) with 2 x 250 bp paired-end reads. Raw sequencing reads were trimmed and filtered based on a Phred score > 20 and minimum fragment length of 50 bp using Solexa QA v3.1.7.1 (Cox et al 2010). Filtered reads were assembled using IDBA-UD v1.1.317 with pre-corrections and genome completeness and contamination were calculated based on lineage-specific marker genes using CheckM v1.1.3.18
DNA from the yeast P. kudriavzevii was extracted using the MasterPure Yeast DNA pu-rification kit (Lucigen). DNA libraries were prepared using the Nanopore Genomic DNA by Ligation kit (SQK-LSK110) and sequencing was performed using the MinION instrument (Oxford Nanopore) and the MinKNOW Sotfware v20.10.3. Fast5 sequencing reads were converted to fastq reads using guppy 3.2.2 (dna_r9.4.1_450bps_hac.cfg). Read quality control was evaluated using NanoFilt v2.7.119 with a Phred score > 20 and minimum fragment length of 50 bp. Filtered sequencing reads were assembled using flye v1.820 and assembly was polished using Medaka v0.12.1 (Oxford Nanopore Technologies Ltd, 2018). Next, assembled contigs were additionally corrected using Illumina reads with Pilon v1.23.21 Assembly statistics were calculated using QUAST v 5.0.222 and genome completeness was evaluated using BUSCO v3 (saccharomycetes_odb10 dataset).23
The sequencing files of this study are deposited in the sequencing read archive (SRA) at National Center for Biotechnology Information (NCBI) under the accession number: PRJNA811803 (Temporal link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA811803?reviewer=jfm49f6ursr8q1qhlgnjlpvhhq) 11803
Covariates
Covariates were measured at the baseline examination and included age (years) and sex. Height (inches) was measured while participants were without shoes and converted into meters. Weight (kilograms), in light clothing was measured with a calibrated digital scale.
Compliance
To monitor compliance, any unused supplements were returned to Solarea Bio and were counted. Two participants who stopped taking supplements early in the course of the study, one due to the start of antibiotics and one due to GI intolerance, were excluded before comparing the two groups on compliance.
Statistical analysis
Baseline characteristics are presented by placebo versus the SBD111 use. Categorical variables are presented as percentages and continuous variables are presented as means and standard deviations (SD). Descriptive variables were compared across the placebo and the SBD111 group using T-tests for continuous variables and Chi-square tests for categorical variables. Primary and secondary outcomes were compared across the two groups using Incidence Rate Ratios (IRR) from the negative binomial regression models or Odds Ratios (OR) from the logistic regression models and 95% confidence intervals (95%CI) using an intention-to-treat approach. Within-person differences in safety and tolerability outcomes (change in the number of AEs, number of GI symptoms, and severity of GI symptoms) between 7 and 28 days were estimated as mean difference and 95%CI using Student’s T-test. Similar analyses were performed for estimating mean difference and 95% confidence intervals for between group changes.
The sample size was determined based on similar studies assessing safety and tolerability of probiotics in healthy adults. The power calculation was based on an assumption of incidence of experiencing gas as a symptom in the active arm and placebo arm of 50% and 5%, respectively. Based on this assumption, 30 participants would have achieved over 80% power to detect such a difference. A nominal two-sided p value of 0.05 was considered statistically significant for all analyses. All analyses were conducted using SAS software version 9.3 (SAS Institute Inc., Cary, NC).
Differential abundant taxa and metabolic pathways between placebo and SBD111 groups at different time points were identified by Linear Discriminant Analysis (LDA) Effect Size v1.1.1 (LEfSe).24 Significance was considered when the logarithmic LEfSe score for discriminative features was 2.0 and P-value < 0.05. Beta diversity was calculated using Bray–Curtis dissimilarity at the species level using the vegan package (version 2.5-4)25 in R. Visualization of microbiome data was performed using ggplot2.26
Study inclusion
A convenience sample of 69 individuals were pre-screened on phone; of these, 33 individuals were deemed eligible to participate in this study (Supplementary Figure S1). One individual refused to participate in the study and 32 participants provided informed consent and were enrolled in the study. During the 28-day follow-up, one participant stopped taking the study product due to consistent GI adverse events and one participant was deemed ineligible when the participant started taking antibiotics. Both participants were in the placebo group. However, both participants were asked to provide stool samples at 28-day and 56-day fol-low-ups. All the 32 participants returned the 28-day stool sample. One stool sample from the 56-day follow-up was lost in transit, resulting in 31 stool samples that were returned at the 56-day follow-up.
Study compliance
Compliance as assessed by counting the number of returned pills [SBD111: 1.2 (SD: 1.7) pills vs. placebo: 4.2 (SD: 6.3) pills, P=0.14] as well as number of compliance days, [SBD111: 27.4 (SD: 0.8) days vs. placebo: 25.8 (SD: 3.8) days, P=0.16] which were not significantly different between SBD111 users compared to placebo users (Table S1).
Baseline characteristics
At baseline, the mean age was 45 (SD: 18) years in the placebo group (n=15, 5M, 10F) and 42 (SD: 15) years in the SBD111 group (n=17, 5M, 12F). The two groups had similar mean BMI, systolic, and diastolic blood pressure at baseline (Table 1).
|
Participant characteristics |
Mean (SD) or n (%) |
|
|
|
Placebo (N=15) |
SBD111 (N=17) |
P Value |
|
|
Sex, n (%) |
|
||
|
Male |
5 (33.3) |
5 (29.4) |
0.81 |
|
Female |
10 (66.7) |
12 (70.6) |
|
|
Age, years |
45 (18) |
42 (14) |
0.6 |
|
BMI, kg/m2 |
25.9 (4.3) |
24.7 (2.7) |
0.33 |
|
Seated SBP, mmHg |
127.7 (14.4) |
123.1 (16.0) |
0.4 |
|
Seated DBP, mmHg |
81.1 (8.7) |
78.4 (9.1) |
0.41 |
|
White Blood Cell |
6.6 (1.7) |
6.0 (1.1) |
0.24 |
|
Hemoglobin |
13.6 (1.9) |
13.5 (1.2) |
0.9 |
|
Platelet Count |
271.9 (67.2) |
246.5 (48.2) |
0.22 |
|
Aspartate Aminotransferase |
20.9 (7.8) |
23.0 (4.7) |
0.35 |
|
Alanine Transaminase |
29.7 (13.1) |
29.9 (7.9) |
0.95 |
|
Compliance, excluding early withdrawals1 — mean (SD) |
|||
|
Number of capsules returned2 |
|||
|
Actual pill count |
4.2 (6.3) |
1.2 (1.7) |
0.14 |
|
Number of days of compliance |
25.8 (3.8) |
27.4 (0.8) |
0.16 |
Table 1 Comparison of demographic variables across placebo and SBD111 users
1Two participants, both in the placebo group, stopped taking the supplement early in the course of the study, one due to the start of antibiotics and one due to GI intolerance.
2Three participants, one in the placebo group, two in the SBD111 group, did not return pill bottles and were missing data on pill count.
Safety and tolerability
During the period of SBD11 or placebo administration, there were no significant differences in the mean number or severity of adverse events, and all were mild or moderate (Table 2). However, the number of AEs that were possibly related to the study were significantly higher in the SBD111 group versus the placebo group (P=0.03). Similarly, no significant differences were seen in the number, frequency or severity of GI symptoms (Table 3).
|
Study outcomes |
Mean (SD) unless otherwise noted |
IRR or OR (95%CI) |
||||
|
Placebo (N=15) |
SBD111 (N=17) |
P Value |
Placebo vs SBD1111 |
|
P Value |
|
|
Total number of AEs |
1.1 (1.5) |
1.8 (1.9) |
0.22 |
1.71 (0.71, 4.10) |
0.23 |
|
|
Number of AEs related to study, n (%) |
0.08 |
|||||
|
Not related |
2 (12.5) |
5 (16.7) |
2.21 (0.42, 11.69) |
0.35 |
||
|
Unlikely related |
5 (31.3) |
6 (20.0) |
1.06 (0.17, 6.60) |
0.95 |
||
|
Possibly related |
5 (31.3) |
18 (60.0) |
3.18 (1.10, 9.17) |
0.03 |
||
|
Probably related |
4 (25.0) |
1 (3.3) |
0.44 (0.05, 4.06) |
0.47 |
||
|
Definitely related |
0 (0) |
0 (0) |
0 vs. 0 |
- |
||
|
Number of AEs by days |
||||||
|
7 |
0. 7 (1.2) |
1.1 (1.1) |
0.35 |
1.59 (0.62, 4.06) |
0.33 |
|
|
14 |
0. 9 (1.4) |
1.2 (1.1) |
0.6 |
1.26 (0.54, 2.92) |
0.59 |
|
|
21 |
1.0 (1.5) |
1.4 (1.4) |
0.57 |
1.27 (0.55, 2.94) |
0.58 |
|
|
28 |
1.1 (1.5) |
1.6 (1.7) |
0.31 |
1.54 (0.66, 3.62) |
0.32 |
|
|
Severity of AEs, n (%) |
||||||
|
Mild |
11 (68.8) |
23 (74.2) |
0.69 |
1.84 (0.76, 4.50) |
0.18 |
|
|
Moderate |
5 (31.3) |
8 (25.8) |
1.41 (0.19, 10.32) |
0.73 |
||
|
Severe/ life threatening |
0 (0) |
0 (0) |
0 vs. 0 |
- |
||
|
Number of serious adverse events (SAEs) |
0 (0) |
0 (0) |
- |
0 vs. 0 |
- |
|
Table 2 Comparison of safety and tolerability outcome variables across placebo and SBD111 users
1Incidence Rate Ratio (IRR) from negative binomial regression models or odds ratio (OR) from logistic regression models comparing placebo to SBD111 users, where placebo is the reference group. These models were run for all outcome variables except for frequency of GI symptoms.
|
Study outcomes |
Mean (SD) unless otherwise notes |
IRR or OR (95%CI) |
|||
|
|
Placebo (N=15) |
SBD111 (N=17) |
P Value |
Placebo vs SBD1111 |
P Value |
|
GI symptoms, n (%) |
5 (33.3) |
11 (64.7) |
0.08 |
3.67 (0.85, 15.84) |
0.08 |
|
Total GI symptoms |
1.7 (4.2) |
2.5 (3.7) |
0.57 |
1.46 (0.42, 5.13) |
0.56 |
|
New GI symptoms by day |
|||||
|
7 |
0.4 (1.4) |
0 (0) |
0.34 |
--2 |
- |
|
14 |
0.7 (1.8) |
0.1 (0.4) |
0.29 |
0.19 (0.01, 2.56) |
0.21 |
|
21 |
0.1 (0.3) |
0.2 (0.8) |
0.54 |
3.00 (0.09, 102.99) |
0.54 |
|
28 |
0 (0) |
0.4 (0.8) |
0.08 |
--3 |
- |
|
Cumulative GI symptoms by day |
|||||
|
0-7 |
1.1 (2.7) |
1.5 (1.6) |
0.61 |
1.38 (0.40, 4.71) |
0.61 |
|
14 |
1.7 (4.2) |
1.6 (1.7) |
0.95 |
0.95 (0.28, 3.26) |
0.94 |
|
21 |
1.7 (4.2) |
1.8 (2.2) |
0.98 |
1.02 (0.29, 3.59) |
0.98 |
|
28 |
1.7 (4.2) |
2.1 (2.8) |
0.76 |
1.22 (0.34, 4.44) |
0.76 |
|
Frequency of GI symptoms, n (%) |
|||||
|
0 |
10 (66.7) |
6 (35.3) |
0.49 |
||
|
1 |
2 (13.3) |
3 (17.6) |
|||
|
2 |
0 (0) |
2 (11.8) |
|||
|
3 |
1 (6.7) |
2 (11.8) |
|||
|
>4 |
2 (13.4) |
4 (23.6) |
|||
|
GI symptoms by severity |
|||||
|
Mild |
1.2 (2.5) |
1.9 (2.9) |
0.49 |
1.57 (0.42, 5.81) |
0.5 |
|
Moderate |
0.5 (1.8) |
0.2 (0.6) |
0.55 |
0.44 (0.04, 4.35) |
0.48 |
|
Severe/life threatening |
0 (0) |
0 (0) |
- |
0 vs. 0 |
- |
|
Duration of side effects, days |
3.4 (3.7) |
3.0 (4.8) |
0.75 |
-0.44 (-3.07, 2.19) |
0.74 |
Table 3 Comparison of safety and tolerability related to gastrointestinal symptoms (GI) across placebo and SBD111 users
1Incidence Rate Ratio (IRR) from negative binomial regression models or odds ratio (OR) from logistic regression models comparing placebo to SBD111 users, where placebo is the reference group. These models were run for all outcome variables except for frequency of GI symptoms.
There were no significant differences between the two groups in the change in the number of AEs, change in GI symptoms, and change in the severity of GI symptoms calculated within each group from day 7 to day 28 (Table 4).
|
Study outcomes |
Mean difference (95% CI) within group change from day 7 to day 28 |
Mean difference (95%CI) between group change1 |
P Value |
|
|
Placebo (N=15) |
SBD111 (N=17) |
Placebo (Ref) vs. SBD111 |
||
|
Change in the number of AEs |
0.4 |
0.59 |
0.19 |
0.41 |
|
(0.05, 0.75) |
(0.22, 0.95) |
(-0.30, 0.68) |
||
|
Change in the number of GI symptoms |
0.67 |
0.65 |
-0.02 |
0.97 |
|
(-0.26, 1.60) |
(-0.10, 1.40) |
(-1.15, 1.11) |
||
|
Change in severity of GI symptoms |
||||
|
Mild |
0.33 |
0.59 |
0.25 |
0.55 |
|
(-0.07, 0.73) |
(-0.16 1.34) |
(-0.60, 1.11) |
||
|
Moderate |
0.33 |
0.06 |
-0.27 |
0.34 |
|
(-0.25 0.91) |
(-0.07, 0.18) |
(-0.86, 0.31) |
||
|
Severe/very severe |
0 vs. 0 |
0 vs. 0 |
- |
- |
Table 4 Within-person differences in safety and tolerability outcomes from 7 days to 28 days across the placebo versus SBD111 group
1Mean difference between group change was calculated as mean values in SBD111 - mean values in the placebo group.
Analyses of the gut microbiome
Shotgun metagenomic analyses of the gut microbiomes of participants revealed no significant differences in alpha diversity indexes (Shannon diversity and microbial richness) between the two groups at baseline, after 28-days of administration, and after 56 days (Mann-Whitney test, P-value < 0.05) (Figure 1). Similarly, there was no difference in beta diversity between the two groups at baseline or over time (Supplementary Figure S2).
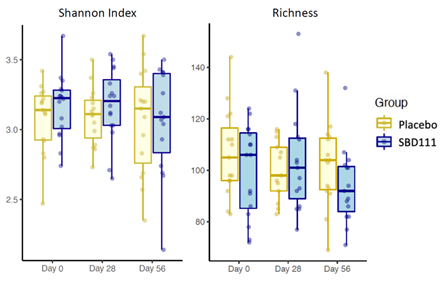
Figure 1 Comparison of alpha diversity indexes between placebo and SBD111 groups at different time points. (Mann-Whitney test, *P-value < 0.05, **P-value < 0.01, comparison between placebo vs. group for each time point).
SBD111 strains were not detected at baseline in either placebo or SBD111 groups except for one participant with L. plantarum, one with Leuc. mesenteroides, and another one with P. kudriavzevii in the product group (Figure 2). After 28-days of administration, there was a variable increase in abundance of all SBD111 strains in the product group that later decreased and became not detectable by 28 days after stopping the intervention. One participant from the placebo group showed an abundance of Leuc. mesenteroides at day 56.

Figure 2 Detection of SBD111 strains in the gut metagenome of participants. Statistical significance of the relative abundance of L. brevis, L. plantarum and L. mesenteroides was evaluated between placebo and product groups for each time point using a LDA Effect Size (LefSe) analysis with P-value of < 0.05 and a LDA score ≥ 2.0 considered significant. Mann-Whitney test was performed to compare the ITS coverage of P. kudriavzevii between groups. *P-value < 0.05, **P-value < 0.01.
Linear Discriminate Analysis (LDA) revealed that after 28 days of test product administration, there were 29 pathways with an increased abundance in the SBD111 group and 9 pathways with a decreased abundance in the placebo group (Supplementary Figure S3). Some abundant pathways at baseline in the SBD111 group were maintained at higher abundance than in the placebo group at day 28 including biotin biosynthesis, 5-oxo-L-proline metabolism, and fatty acid & beta-oxidation IV. Moreover, other pathways were enriched in the SBD111 group including those related to menaquinone (vitamin K2) production (super pathways of menaquinol-6, -9, and -10) (Figure 3). During the washout period, the vitamin K2 pathway enrichment observed during the intervention period was no longer present, but eight other metabolic pathways increased in abundance in the SBD111 group including those involved in lipid biosynthesis, peptidoglycan maturation and 8 decreased in the placebo group.

Figure 3 Metabolic pathways associated with vitamin K biosynthesis with differential abundance between placebo and SBD111 groups at day 28 identified by LDA Effect Size (LefSe) analysis (P-value of < 0.05 and an LDA score ≥ 2.0). PWY-5845: superpathway of menaquinol-9 biosynthesis, PWY-5850: superpathway of menaquinol-6 biosynthesis, PWY-5860: superpathway of demethylmenaquinol-6 biosynthesis I, PWY-5862: superpathway of demethylmenaquinol-9 biosynthesis, PWY-5896: superpathway of menaquinol-10 biosynthesis.
Further, after 28-days of test product administration, fragment recruitment analysis re-ported the genomic signal of SBD111 strains in the gut metagenomes of most participants receiving SBD111, but none of those receiving placebo (Supplementary Figure S4). The genomic signal was not detectable after the wash out period (day 56) in the SBD111 group.
In this study, we evaluated safety and tolerability of SBD111, an optimized synbiotic medical food designed for the dietary management of the metabolic processes of osteopenia and osteoporosis. We found that the two study groups were not significantly different in mean number or severity of AEs. Similarly, no significant differences were seen in the number, frequency or severity of GI symptoms or compliance across the two groups. The only statistically significant difference was that the number of AEs that were possibly related to the study were significantly higher in the SBD111 group versus the placebo group. However, two participants from the placebo group stopped taking supplements early in the course of the study, one due to the start of antibiotics and one due to GI intolerance.
We further showed that SBD111 strains were able to pass through the intestinal tract, be identified in stool, as well as provide relevant and potentially bone-protective gene functionalities including vitamin K2 production at 28 days. Furthermore, we found that the genomic signal of SBD111 could not be detected after the washout phase at day 56.
Modification of the gut microbiota is an emerging area of research in relation to chronic diseases of aging, including osteoporosis. Experimental studies in animals show that gut microbial changes can affect calcium balance, inflammation, immunity, and bone homeostasis.27-29 Yet, data from human clinical trials are limited. Recent preclinical and clinical studies suggest that targeting specific key taxa related to beneficial effects to the host may be a safe and viable target for the dietary management of skeletal homeostasis. In mice, probiotics were protective against bone loss and improved markers of bone turnover.28,30-34 Postmeno-pausal osteopenic women administered probiotics for 12-months were protected from bone loss compared to placebo.5,6,35 Several mechanisms underlying the observed skeletal effects of probiotics have been suggested:
These previous pre-clinical and early human studies make a compelling case for the development of an optimal probiotic and prebiotic combination. Our study is one of the first to test such a product for safety and tolerability. We completed our recruitment goals, with relatively high interest and compliance with our study protocol. Only two adults (both in the placebo group) discontinued the supplements, one due to use of antibiotics and the other due to GI intolerance. Study compliance was quite good and did not differ across the two groups. Our GI adverse events were ascertained daily during the early period to provide a thorough assessment of side effects.
Our study has limitations. First, we were underpowered to detect small differences in adverse events. Second, we only measured stool microbiota at baseline, after 28 days of ad-ministration, and after an additional 28 day washout period. To understand the dynamics of product detection in stool upon the start of the intervention and the washout period will require more frequent sampling and higher sample size. Despite these limitations, our study has many strengths. We executed a rigorous blinded, randomized, controlled design to test the safety and tolerability of SBD111 in humans. The concept of combining an optimal formulation of prebiotics and probiotics in a single formulation is a significant development over administering single probiotics without attention to the accompanying need for a fiber substrate. Shotgun metagenomic analyses provided important insight on the genomic detection of SBD111 strains during the study in addition to the identification of metabolic pathways associated with vitamin K2 biosynthesis. We utilized a classification system for AEs such that both total as well as GI specific AEs were recorded. GI tolerability was measured by a validated questionnaire.14 Inclusion of a wash out phase provided additional information on safety features of SBD111.
In conclusion, this study demonstrates the safety and tolerability of SBD111 in adult men and women. Our study suggests that SBD111 strains are able to pass through the intestinal tract, be identified in stool, as well as provide relevant and potentially bone-protective gene functionalities including vitamin K2 production. Therefore, future studies should test whether SBD111 will improve bone health in a larger clinical trial by measuring bone-protective efficacy in a relevant target population.
The authors’ responsibilities were as follows-SS, EMS, and DPK designed the research and acquired funding; DPK served as the study physician; DC provided project management support and collected data; SS, MJSG, and EMS analyzed data with critical input from all co-authors; SS, EMS, and MJSG drafted the manuscript with editing by all authors; SS had primary responsibility for final content; and all authors read and agreed to the published version of the manuscript.
This research was funded by Solarea Bio Inc. SS was in part supported by the National Institute on Aging support of the Boston Claude D. Pepper Center Older American In-dependence Centers (OAIC; 1P30AG031679) and Peter and Barbara Sidel Fund. Role of funding agency: Solarea Bio Inc. participated in study design, data analysis, interpretation, and drafting of this manuscript.
This study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Advarra (protocol code Pro00054643 and 25th June 2021).
Informed consent was obtained from all subjects involved in the study.
Data described in the manuscript, code book, and analytic code will be made available upon request pending application to and approval by Solarea Bio Inc.
We are indebted to the study participants for their sustained commitment to the Solarea clinical trial. We thank Dr. D. Davidson Easson, Jr. from Solarea Bio Inc. for his instrumental role in the production and quality assurance of both SBD111 and placebo test articles; Ms. Nessa Steinberg from the Marcus Institute, Hebrew SeniorLife for providing part of the administrative support; and Mr. Daniel Habtemariam from the Marcus Institute, Hebrew SeniorLife for his programming efforts.
Dr. Sahni reports institutional grants from Dairy Management Inc. and Solarea Bio Inc., has reviewed grants for the American Egg Board’s Egg Nutrition Center and serves on a scientific advisory board for the Protein Committee, the Institute for the Advancement of Food and Nutrition Sciences. Dr. Kiel serves as a consultant to Solarea Bio Inc., Reneo, and Pfizer, and receives grant support from Amgen, Solarea Bio Inc., and Radius Health, and royalties for publication in UpToDate by Wolters Kluwer. Other authors have no conflicts of interest to report..
Study procedures |
Prescreening |
Screening |
Baseline |
D1-6 |
D7 |
D14 |
D21 |
D28 |
D56 |
Demographics |
X |
||||||||
Age |
X |
||||||||
Medical History |
X |
X |
|||||||
Medications & Supplements |
X |
X |
|||||||
Height |
X |
X |
|||||||
Weight |
X |
X |
|||||||
BMI Calculation |
X |
X |
|||||||
Blood Pressure |
X |
X |
|||||||
Urine Sample |
X |
||||||||
White Blood Cell Count |
X |
||||||||
Neutrophil Count |
X |
||||||||
Platelet Count |
X |
||||||||
Hemoglobin |
X |
||||||||
Aspartate Aminotransferase |
X |
||||||||
Alanine Transaminase |
X |
||||||||
Total Bilirubin |
X |
||||||||
Randomization |
X |
||||||||
Instruction Distribution |
X |
||||||||
Adherence Assessment |
X |
X |
X |
X |
X |
||||
Stool Collection |
X |
X |
X |
||||||
AE Reporting |
X |
X |
X |
X |
X |
X |
|||
GITQ |
|
|
X |
X |
X |
X |
X |
X |
|
Table S1 Schedule of study assessments
Material S2 Eligibility criteria
Inclusion
Eligible participants were aged 18-70 years, have a Body Mass Index between 18.5 and 35 kg/m2, systolic blood pressure ≤ 155 mm Hg, diastolic blood pressure ≤ 95 mm Hg.
Exclusion
Exclusion criteria included:
|
Ingredient |
Dose in Each Capsule (CFUs) |
Daily Dose in Two Capsules (CFUs) |
Ingredient in Each Capsule (mg) |
|
Lactobacillus brevis SBS4254 |
9.6x109 |
1.9 x1010 |
15-30 |
|
Lactobacillus plantarum SBS4260 |
9.6x109 |
1.9 x1010 |
15-30 |
|
Leuconostoc mesenteroides SBS4255 |
9.6x109 |
1.9 x1010 |
15-30 |
|
Pichia kudriavzevii SBS4263 |
1.6x109 |
3.2 x109 |
75-160 |
|
Oligofructose |
NA |
NA |
125-235 |
|
Dried, ground blueberry powder |
NA |
NA |
125-235 |
|
Magnesium stearate |
NA |
NA |
5-10 |
|
Silicon dioxide |
NA |
NA |
5-10 |
|
Total |
3.0x1010 |
6.1x1010 |
500-550 |
Table S3 Ingredients in Solarea Bio’s SBD111 Medical Food
Abbreviations: CFU, colony forming units; NA, not applicable, not part of microorganism dose
|
Frequency of individual GI symptoms from GITQ, n (%) |
Mean (SD) or n (%) |
|
IRR or OR (95%CI) |
||
|
Placebo (N=15) |
SBD111 (N=17) |
P Value |
Placebo vs SBD1111 |
P Value |
|
|
Bloating |
2 (13.3) |
4 (23.5) |
0.46 |
2.00 (0.31, 12.89) |
0.47 |
|
Gas |
2 (13.3) |
8 (47.1) |
0.04 |
5.78 (0.99, 33.82) |
0.05 |
|
Intestinal Rumbling |
3 (20.0) |
7 (41.2) |
0.2 |
2.80 (0.57, 13.75) |
0.2 |
|
Blood in Stool |
0 (0) |
0 (0) |
- |
0 vs. 0 |
- |
|
Abdominal Cramps or Pain |
4 (26.7) |
3 (17.6) |
0.54 |
0.59 (0.11, 3.20) |
0.54 |
|
Nausea |
1 (6.7) |
1 (5.9) |
0.93 |
0.88 (0.05, 15.33) |
0.93 |
|
Vomiting |
0 (0) |
0 (0) |
- |
0 vs. 0 |
- |
|
Loss of Appetite |
1 (6.7) |
1 (5.9) |
0.93 |
0.88 (0.05, 15.33) |
0.93 |
|
Abnormal Taste |
0 (0) |
0 (0) |
- |
0 vs. 0 |
- |
|
Heartburn |
1 (6.7) |
2 (11.8) |
0.62 |
1.87 (0.15, 22.94) |
0.63 |
|
Constipation |
0 (0) |
3 (17.6) |
0.09 |
--2 |
- |
Table S4 Comparison of safety and tolerability related to gastrointestinal (GI) symptoms between placebo and SBD111 users
1Incidence Rate Ratio (IRR) from negative binomial regression models or odds ratio (OR) from logistic regression models comparing placebo to SBD111 users, where placebo is the reference group; 2Infinite odds ratio, 0 odds in the placebo group.
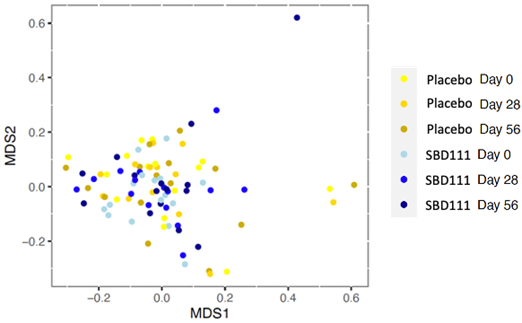
Figure S2 Non-metric Multidimensional Scaling (NMDS) Plot of Beta diversity with Bray-Curtis Dissimilarity Distances at the species level. PERMANOVA analyses did not show significance between groups or time points (P-value=1).
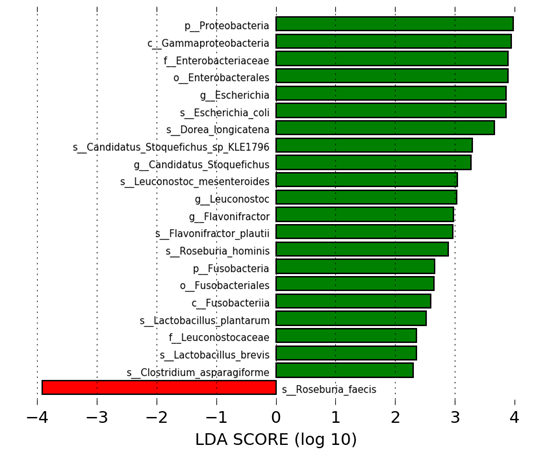
Figure S3 Microbial taxa and metabolic pathways with differential abundance between placebo and SBD111 groups at 28-days identified by Linear Discriminant Analysis (LDA) Effect Size (LEfSe) (p-value of < 0.05 and a LDA score ≥ 2.0). The LDA score corresponds to the degree of difference in the magnitude of relative abundances of features that differ between the two groups.
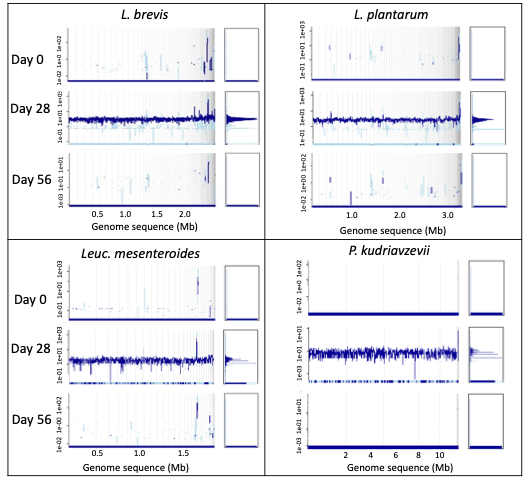
Figure S4 Detection of SBD111 strains in the gut metagenome at day 28 of participants in the SBD111 group. Fragment recruitment plot of the metagenomic sample from one participant taken at each time point. The plot shows the sequencing depth values in a logarithm scale of metagenomics reads that were mapped onto each SBD111 genome sequence. The dark blue peak corresponds to the average coverage of reads that were mapped with ≥95% nucleotide identity. The light blue represents the average coverage of reads with <95% nucleotide identity. Fragment recruitment plots were built using the enveomics.R package v1.4.1 from the Enveomics collection.27

©2023 Sahni, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.