Journal of
eISSN: 2373-437X


Research Article Volume 8 Issue 1
1College of Landscape Architecture, Jiangsu Vocational College of Agriculture and Forestry, China
2Department of Plant Pathology, Nanjing Agricultural University, China
Correspondence: Jia-Jia Chen, Department of Plant Pathology, Nanjing Agricultural University, Nanjing 210095, China
Received: December 16, 2019 | Published: January 21, 2020
Citation: Yu J, Zhou YH, Yang HJ, et al. Pythium glomeratum and Py. nodosum, two new records from China. J Microbiol Exp. 2020;8(1):24-28. DOI: 10.15406/jmen.2020.08.00281
Two oomycetes, Pythium glomeratum and Py. nodosum were found in southern China, and they are newly recorded in China. These two species were both isolated from roots of soybean. Py. glomeratum is characterized by absence of zoosporangia and zoospores, and slow growth rate. And Py. nodosum is identified from other Pythium species by its smooth oogonia which are crowded with different antheridial branches making a complicated knot, and aplerotic oospores. Illustrations and descriptions of the two new records are provided based on the materials from China.
Keywords: Cox1, ITS, Oomycota, Pythium glomeratum, Pythium nodosum
BI, bayesian inference; BPP, bayesian posterior probabilities; BT, bootstrap; CI, consistency index; CMA, corn meal agar; Cox1, cytochrome c oxidase subunit 1; CTAB, cetyl trimethylammonium bromide; GTR, general time reversible; HI, homoplasy index; ITS, the internal transcribed spacer; MP, maximum parsimony; MPT, maximum parsimonious tree; NJAU, the College of Plant Protection, Nanjing Agricultural University; PCA, potato carrot agar; PCR, the polymerase chain reaction; RC, rescaled consistency index; RI, retention index; TL, descriptive tree statistics tree length
The genus Pythium Pringsheim1 was typified by Py. monospermum Pringsh. The principal characteristics of this genus include a combination of characterized by hyaline and coenocytic hyphae without septa, various shaped sporangia, and the development of zoospores in a vesicle which is formed at the tip of a discharge tube derived from a sporangium.2 Pythium spp. are cosmopolitan and represent a range of functional groups, such as saprophytes in natural environments, plant and animal pathogens, and biological control agents protecting against pathogenic fungi.3 Following recent taxonomic revisions4,5 and discoveries,6-8 more than 140 species are currently recorded in the genus Pythium.8 During studies on the occurrence and diversity of Pythium in China, two new Chinese record of Pythium were identified from our isolates based on morphological characters and molecular phylogenetic analyses of ITS regions of the ribosomal RNA and mitochondrial Cox1 sequence data. These two species are described in this work.
Isolates
The cultures (Chen 276, 409 & 412) of Pythium species were isolated from roots of soybean in Beijing and Jiangsu provinces in China. The isolation procedure followed the method described by Benard & Punja.9 Pieces of tissue 5–10 mm were cut from the roots, washed in tap water and superficially dried on a paper towel, and plated on CMA containing rifampicin (50mg/L), phenamacril (5mg/L), ampicillin (50mg/L), and pentachloronitrobenzene (50mg/L) and incubated at 25°C for 2–3 d. When mycelial growth was observed, purification was carried out by cutting a small piece of medium with mycelia at the edge of a colony, and transferring the cutted part into the new medium plates.
Morphology and growth rate
The studied cultures were deposited in the herbaria of NJAU. The purified isolates were grown on CMA for morphological studies. Isolates were transferred to sterilized distilled water for sporulation. Fifty measurements were taken for each morphological feature, such as sporangia, oogonia and oospores. The cardinal temperatures were examined on PCA according to the method of van der Plaäts-Niterink,2 and growth rates were measured at 24 h incubation. Each isolate was incubated at 5–40°C with intervals of 5°C on PCA media. When no growth was observed, the intervals were reduced from 5 to 2 or 1°C and the culture was returned to room temperature to check the revival of the growth.10
Molecular phylogeny
DNA extraction, amplification, sequencing and sequence alignment
A CTAB rapid plant genome extraction kit (Demeter Biotechnologies Co., Ltd, Beijing) was used to extract total genomic DNA from purified isolates, and performed PCR according to the manufacturer’s instructions with some modifications.7
The ITS region was amplified with the primers: ITS4 and ITS5.11 The Cox1 gene was amplified with the primers: OomCoxI-Levlo (CYTCHGGRTGWCCRAAAAACCAAA) and OomCoxI-Levup (TCAWCWMGATGGCTTTTTTCAAC).12 The PCR procedure for ITS was as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 54°C for 45 s and 72°C for 1 min, and a final extension of 72°C for 10 min. The PCR procedure for Cox1 was as follows: initial denaturation at 94°C for 2–5 min, followed by 35 cycles at 94°C for 30 s, 52°C for 30 s and 72°C for 1–2 min, and a final extension of 72°C for 5–10 min.13 The PCR products were purified and sequenced in Genscript company (Nanjing, China) with the same primers.
Sequences generated in this study were aligned with additional Pythium sequences downloaded from GenBank (Table 1) using ClustalX14 and manually adjusted in BioEdit.15 Sequence alignment was deposited at TreeBase (http://purl.org/phylo/treebase; submission ID S25483).
Species name |
Isolate no. |
Geographic origin |
GenBank accession no. |
|
ITS rDNA |
Cox1 mtDNA |
|||
Phytopythium ostracodes |
CBS 768.73 |
Spain |
HQ643395 |
HQ708442 |
Pythium baisense |
HMAS 242232 |
China |
FR775440 |
FR774198 |
Py. breve |
HMAS 242231 |
China |
FR751317 |
FR774196 |
Py. buismaniae |
CBS 288.31 |
Netherlands |
HQ643479 |
HQ708526 |
Py. cystogenes |
CBS 675.85 |
Netherlands |
HQ643518 |
HQ708564 |
Py. glomeratum |
CBS 122651 |
France |
HQ643541 |
HQ708585 |
Py. glomeratum |
Chen 276 |
China |
MN732893 |
MN756479 |
Py. heterothallicum |
CBS 450.67 |
Canada |
HQ643553 |
HQ708597 |
Py. sp. jasmonium |
DAOM 229150 |
USA |
HQ643670 |
HQ708714 |
Py. mastophorum |
CBS 375.72 |
United Kingdom |
HQ643691 |
HQ708735 |
Py. megalacanthum |
DAOM 229154 |
Germany |
HQ643693 |
HQ708737 |
Py. nodosum |
CBS 102274 |
France |
HQ643709 |
HQ708753 |
Py. nodosum |
Chen 409 |
China |
MN732894 |
MN756480 |
Py. nodosum |
Chen 412 |
China |
MN732895 |
MN756481 |
Py. nunn |
CBS 808.96 |
USA |
HQ643711 |
HQ708755 |
Py. orthogonon |
CBS 376.72 |
Lebanon |
HQ643723 |
HQ708764 |
Py. perplexum |
CBS 674.85 |
Netherlands |
HQ643744 |
HQ708785 |
Py. polymastum |
CBS 811.70 |
Netherlands |
HQ643752 |
HQ708793 |
Py. splendens |
CBS 462.48 |
USA |
HQ643795 |
HQ708836 |
Py. ultimum var. sporangiiferum |
CBS 219.65 |
USA |
HQ643879 |
HQ708920 |
Py. ultimum var. ultimum |
CBS398.51 |
Netherlands |
HQ643865 |
HQ708906 |
Py. uncinulatum |
CBS 518.77 |
Netherlands |
HQ643944 |
HQ708985 |
Table 1 A list of species, cultures, and GenBank accession numbers of sequences used in this study
New sequences are shown in bold.
Phylogenetic analyses
Phylogenetic analysis was done as in Chen & Cui.16 MP analysis was applied to the combined dataset of ITS-Cox1 sequences. Phytopythium ostracodes (Drechsler) Abad, de Cock, Bala, Robideau, A.M. Lodhi & Lévesque was used as an outgroup. The tree construction procedure was performed in PAUP* version 4.0b10.17 All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a BT analysis with 1000 replicates.18 TL, CI, RI, RC, and HI were calculated for each MPT generated. Phylogenetic trees were visualized using Treeview.19
MrModeltest2.320 was used to determine the best-fit evolution model for BI. BI of the dataset was calculated with MrBayes3.1.2 (Ronquist and Huelsenbeck 2003) with a GTR model of DNA substitution and an inverse gamma distribution rate variation across sites. Four Markov chains were run for 2 runs from random starting trees for 2 million generations of the two combined datasets, and trees were sampled every 100 generations. The burn-in was set to discard the first 25% of the trees. A majority rule consensus tree of all remaining trees was calculated. Branches that received bootstrap support for MP and BPP greater than or equal to 75% (MP) and 0.95 (BPP) respectively were considered as significantly supported.
Molecular phylogeny
The combined ITS + Cox1 dataset included sequences from 23 isolates representing 20 taxa. The dataset had an aligned length of 1662 characters, of which 842 characters are constant, 218 are variable and parsimony-uninformative, and 602 are parsimony-informative. MP analysis yielded one equally parsimonious tree (TL=1868, CI=0. 675, RI=0.806, RC=0.543, HI=0.325). The best model for the combined ITS + Cox1 sequences dataset estimated and applied in the BI was GTR+I+G. BI resulted in a similar topology with an average standard deviation of split frequencies = 0.001950 to MP analysis, and thus only the MP tree was provided. Both BT values (≥50%) and BPPs (≥0.95) are shown at the nodes (Figure 1). The Chinese isolates Chen 276, 409 & 412 were identical to the authorized sequences of Pythium glomeratum and Py. nodosum available in GenBank and thus clustered within clades representing Py. glomeratum and Py. nodosum with high supporting values (Figure 1), respectively.
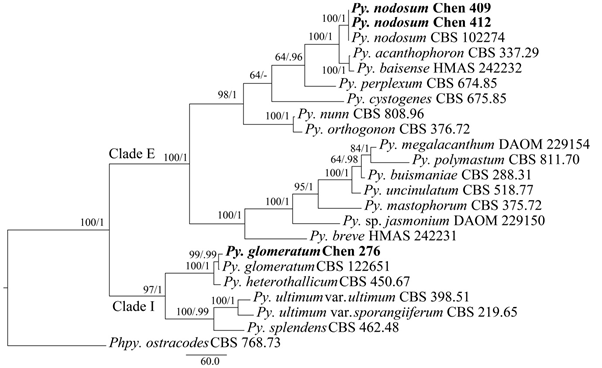
Figure 1 Phylogeny of Pythium glomeratum and Py. nodosum and related species generated by maximum parsimony based on ITS-Cox1 sequence data. Branches are labeled with parsimony bootstrap proportions (before slanting line) high than 50% and Bayesian posterior probabilities (after slanting line) more than 0.95. Pp. refers to Phytopythium, and Py. refers to Pythium.
Taxonomy
Pythium glomeratum B. Paul, FEMS Microbiol. Lett. 225: 49, 2003 Figure 2
Colonies submerged, with radiate pattern on CMA. Average growth rates 7 mm day−1 at 10°C, 10 mm day−1 at 15°C, 12 mm day−1 at 20°C, 14 mm day−1 at 25°C, 10 mm day−1 at 30°C, 6 mm day−1 at 35°C, but when returned to room temperature both of them started to grow again. Cardinal temperatures: minimum 5°C, optimum 25°C, maximum 36°C. Main hyphae hyaline, aseptate, up to 5.0µm wide. Hyphal swellings globose, sub-globose, ovoid to peanut shaped, 5–30 (mean 16.5)μm in diameter. Sporangia and zoospores not observed. Homothallic; oogonia globose, smooth, 17.5–25µm (mean 21µm) in diameter. Antheridia not observed. Oospore one per oogonium, plerotic, globose, 15.5–23.5μm (mean 18.5µm) in diameter, wall up to 0.5–2 (mean 1.6)µm thick.
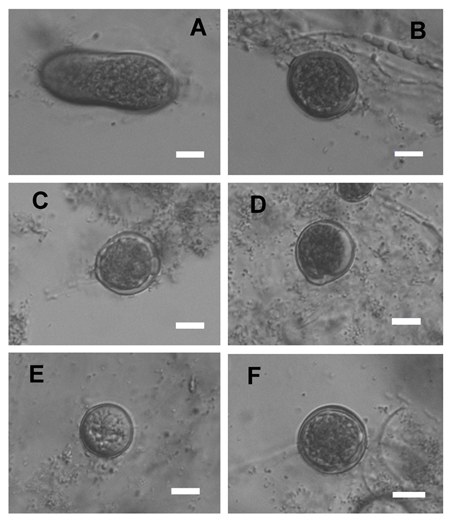
Figure 2 Asexual and sexual reproductive bodies of Pythium glomeratum (Chen 276). A-B. Hyphal swellings. C-F. Globose oogonia. Scale bars A-E=10μm.
Specimen examined: CHINA. Beijing, from Glycine max, 7 Sep 2017, J.J. Chen, Chen 276 (NJAU).
Colonies submerged, with radiate pattern on CMA. Average growth rates 10 mm day−1 at 10°C, 12 mm day−1 at 15°C, 15 mm day−1 at 20°C, 20 mm day−1 at 25°C, 18 mm day−1 at 30°C, 4 mm day−1 at 35°C, but when returned to room temperature both of them started to grow again. Cardinal temperatures: minimum 5°C, optimum 25°C, maximum 36°C. Main hyphae hyaline, aseptate, up to 7.0µm wide. No hyphal swellings. Sporangia globose to sub-globose, 10–27μm (mean 20μm) in diameter. Zoospores formed in sterile water at 25°C; Homothallic; oogonia globose, smooth, terminal, 10–25µm (mean 18.5µm) in diameter. Antheridia monoclinous, one to three per oogonium; antheridial stalks unbranched; antheridial cells elongate, more or less lengthwise applied but crook necked, making narrow apical contact with the oogonium. Oospores aplerotic, globose, 8–21.5 μm (mean 15.5 µm) in diameter, hyaline. Oospore wall thin, 0.5–2µm (mean 1.2µm) thick.
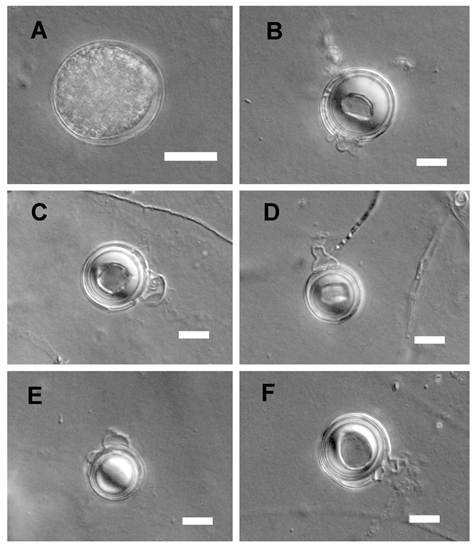
Figure 3 Asexual and sexual reproductive bodies of Pythium nodosum (Chen 409). A. Sub-globose sporangium. B-E. Globose oogonia, aplerotic oospores. Scale bars A-E=10μm.
Specimen examined: CHINA. Jiangsu Province, Nanjing, from Glycine max, 20 June 2017, J.J. Chen, Chen 409 & 412 (NJAU).
In this study, we analyzed the phylogenetics of 14 species of Pythium clade E and five species of Pythium clade I. With the aid of morphology and phylogenetic analyses of the phylogeny of ITS and Cox1 genes, two new Chinese record, Pythium glomeratum and Py. nodosum are described. Pythium glomeratum has a closer relationship with Py. heterothallicum W.A. Campb. & F.F. Hendrix to the ITS and Cox1-based phylogeny (Figure 1). Py. glomeratum resembles Py. heterothallicum in having globose oogonia, but it is distinguished in its absence of zoosporangia and zoospores, slower growth rate and presence of sex organs.22 Pythium nodosum is easily identified by its smooth oogonia which are crowded with different antheridial branches making a complicated knot, and aplerotic oospores. Py. acanthophoron Sideris and Py. baisense Y.Y. Long, J.G. Wei & L.D. Guo resemble Py. nodosum and the combined ITS and Cox1 sequences also suggested a close relationship between the these two species and Py. nodosum within Pythium clade E (Figure 1); however, Py. acanthophoron has ornamented oogonia,2 while Py. baisense is distinguished from Py. nodosum by faster growth rate, and presence of double oospores.10 This study significantly improved our understanding of the oomycetes genera Pythium associated with soybean from China. The two species obtained from this study may potentially be highly valuable. However, because little was known about the role or importance of Pythium spp. in disease on soybean in China. Thus, a further study was undertaken to determine the identity, role, pathogenicity, and virulence of Pythium spp. associated with soybean in the future.
The research was supported by Jiangsu Vocational College of Agriculture and Forestry Research Project (110751168).
Authors declare that there is no conflict of interest.

©2020 Yu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.