Journal of
eISSN: 2373-437X


Research Article Volume 8 Issue 6
Department of Microbiology, Faculty of Science, University of Lagos, Nigeria
Correspondence: Ekpunobi Nzube Favour, Department of Microbiology, Faculty of Science, University of Lagos, Lagos, Nigeria, Tel 08170815462, 07014643819,
Received: November 24, 2020 | Published: December 29, 2020
Citation: Favour EN, Isaac AA. Phenotypic characterization of biofilm formation and efflux pump activity in multi-drug resistant staphylococcus species isolated from asymptomatic students. J Microbiol Exp. 2020;8(6):223-229. DOI: 10.15406/jmen.2020.08.00313
Staphylococcus spp. are one of the major groups of bacterial commensals isolated from skin and mucous membranes. While a variety of Staphylococcal species are present on or in clinically normal individuals, some are also opportunistic pathogens and leading causes of community associated diseases in humans and animals worldwide. The objective of the study was to determine the antimicrobial susceptibility pattern, biofilm formation capability and efflux pump activity in Staphylococcus spp. isolated from nares and armpits of students. A total of 91 swab samples were collected from consenting microbiology students. Characterization of isolates was done using conventional microbiological guidelines yielding 50 Staphylococcal isolates and antimicrobial susceptibility was done following CLSI guidelines. Biofilm formation was detected using the congo red agar method and efflux activity was assayed by a modification of the EtBr cart wheel method using local blue dye. Among the 50 staphylococcal isolates, 7(14%) were coagulase positive while 43(86%) were coagulase negative. Antimicrobial susceptibility pattern of isolates showed resistance of 46(92%) to augmentin, 45(90%) to cloxacillin, 43(86%) to erythromycin, 41(84%) to cefuroxime and 39(78%) to ceftazidime. Resistance to gentamicin and oflaxacin was observed in only 5(10%) of the isolates. Biofilm formation (blackening of medium) was observed in 10(36%) of the 28 multi-drug resistant isolates and 5(83%) of the 6 isolates screened positive for efflux activity (non-retention of blue dye). Biofilm formation and efflux activity have shown to be mechanisms for drug resistance in community-associated staphylococcal isolates with high multi-drug resistance pattern.
Keywords: antimicrobial susceptibility, biofilm formation, efflux pump activity, multi-drug resistance, Staphylococcus spp
Staphylococci are one of the major groups of bacterial commensals isolated from skin, skin glands, and mucous membranes of mammals.1 While a variety of staphylococcal species are present on or in clinically normal individuals, staphylococci are also opportunistic pathogens and leading causes of community-associated disease in humans and animals worldwide.2-4 Being among the microorganisms most resistant to antibiotics, these organisms cause serious infections, whether in hospitalized individuals or healthy individual, emphasizing the importance of epidemiological vigilance in detecting development of resistance in the community.
Methicillin-resistant S. aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci (MRCoNS) are a serious problem for human and animal populations.5 Methicillin-resistant staphylococci (MRS) are present in the human population and may be a reservoir of new or rare strains of MRSA and MRCoNS. Moreover, the importance of carriage of CoNS in the nose, including MRCoNS, both in humans and animals, has not been elucidated.6 In addition, coagulase-negative staphylococci (CoNS) act as reservoirs of resistance genes, although these microorganisms are less virulent. The excessive use of antimicrobial agents and inappropriate empirical treatments has contributed to the growing number of infections by multi-resistant microorganisms in both the community and hospital environment.7 As a result, the treatment of patients with these infections is becoming more complex, greatly increasing the costs of both hospitalization and treatment of these patients in public hospitals.8
In addition to the challenges posed by antimicrobial resistance, the treatment of staphylococcal infections is further complicated by a number of strategies that staphylococci have developed, which enable the bacteria to evade the host immune response and the activity of antimicrobials.9 One important strategy is the formation of biofilms. Bacterial biofilm is a complex community of microorganisms with production of extracellular polysaccharide matrix on damaged tissue and surface of indwelling medical devices. The bacteria encased in biofilms are able to tolerate significantly higher concentrations of antimicrobials and disinfectants than free-floating bacterial cells.9-11 Beenken and co-workers12 revealed a change in the expression of 580 genes (more than 20% of the genome) when using microarrays to study differences between S. aureus cells growing in biofilm and planktonic cultures.
The possession of active multidrug resistance efflux pumps can also mediate antibiotic resistance in bacteria.13 The genetic elements encoding efflux pumps may be encoded on chromosomes and/or plasmids, thus contributing to both intrinsic and acquired resistance respectively.14 Expression of several efflux pumps in a given bacterial species may lead to a broad spectrum of resistance when considering the shared substrates of some multidrug efflux pumps, where one efflux pump may confer resistance to a wide range of antimicrobials.15 This study aims at the determination of antimicrobial resistance pattern, biofilm production and efflux pump activity in Staphylococcus spp., isolated from asymptomatic students.
Study area
Study was conducted at the Department of Microbiology, University of Lagos, Akoka from May to September, 2019. The materials for the collection of samples were shown to the students and an overview of the study was also described to the participants.
Sample collection and processing
Samples were obtained from consenting 400 level students of Microbiology, University of Lagos, Akoka. Nasal and armpit swabs were collected from both males and females aged between 20 and 27 year old. A total of 45 males and 46 females were selected for collection of swabs. Swabs from each student was coded with the letter “N” followed by a numeric number for nasal swab and “A” followed by numeric number for armpit swabs. Also, the male and female students were also noted. Ninety-one (91) swab samples were collected comprising of 62 nares and 29 armpit swabs (Table 1).
Armpit |
Nares |
||
Male ♂ |
Female ♀ |
Male ♂ |
Female ♀ |
16 |
13 |
29 |
33 |
Table 1 Distribution of samples collected from students
With a single swab, a single sample was collected from either the armpits or nasal sites, from each consenting participants. Health care facility exposure, antibiotic usage 2 weeks prior to sample collection and use of deodorants were used as exclusion criteria. The cotton tip of each swab stick was moistened in sterile normal saline and gently rolled over the nares and armpits for about 10 seconds and properly labeled.
Identification of isolates
Each swab sample was immediately streaked on mannitol salt agar (MSA) plates. The plates were incubated at 37oC aerobically and examined for growth after 24 hours. Typical colonies were picked aseptically and purified by sub- culturing onto fresh MSA plates. Staphylococcus spp. was identified on the basis of microscopy, gram stain reaction, catalase test, heamolysis test, coagulase production, and mannitol fermentation.
Antibiotics susceptibility test
Antimicrobial susceptibilities of staphylococci to commonly used drugs were determined by the Kirby–Bauer disc diffusion test on Mueller–Hinton agar and interpreted as recommended. Abtek multiple sensitivity discs, Vancomycin (30μg) and Novobiocin (30μg) single discs were used and the results were interpreted using the clinical and laboratory standards institute guidelines.16 Briefly, isolates were grown to mid-exponential phase, adjusted to 0.5 McFarland opacity tube, and overlaid on Mueller–Hinton agar. The multiple discs comprised the following antibiotics: Ceftazidime (30μg), Cefuroxime (30μg), Gentamicin (10μg), Ceftriaxone (30μg), Erythromycin (5μg), Cloxacillin (5μg), Oflaxacin (5μg), Augmentin (30μg). Duplicate plates were made for each isolates and incubation was done at 37ºC for 24 hours and after incubation, the inhibition zone diameters (IZD) were read using a scale rule and results recorded and compared with the CLSI, 2018 standard. Staphylococcus aureus ATCC 29213 was used as control organism.
Assay for biofilm formation
The Congo red Agar method (CRA) as described by Freeman et al.,17 was used for screening biofilm formation by the isolates. The method requires the use of specifically prepared solid medium–brain heart infusion broth (BHI) supplemented with 5% sucrose and congo red. Congo red was prepared as concentrated aqueous solution and autoclaved at 121oC for 15 minutes, separately from other medium constituent and was then added when agar had cooled. Plates were inoculated and incubated aerobically for 24 hours at 37oC.
Determination of efflux pump activity
The efflux pump activity of the multidrug resistant isolates was determined by modification to the method of the ethidium bromide (EtBr) cartwheel test described by Anbazhagan et al.18 Briefly, Mueller- Hinton agar plates containing 1μg/ml, 1.5μg/ml, 2μg/ml of commercial blue dye were prepared on the same day of the experiment. Each of the Isolates was streaked as cart wheel pattern on commercial blue plates. The plates were wrapped in aluminum foil to protect from light and incubated overnight at 37oC. Thereafter, the plates were viewed under UV light (a transilluminator was used). The isolates were observed for presence or absence of fluorescence.
From the 91 swabs that were plated for the study, there was an appreciable bacterial growth in all but one of the swabs on mannitol salt agar (MSA). All isolates were Gram positive with 7 (7.78%) of the isolates being coagulase positive and 83 (92.22%) being coagulase negative. Microscopy also showed that out of the 90 isolates 30 (33.33%) were bacilli, 10 (11.1%) were cocci in pairs while 50 (55.56%) were cocci in clusters, suspected to be Staphylococcus spp. The 50 suspected Staphylococcus isolates were subjected to further biochemical tests and confirmed as Staphylococcus spp.
Antimicrobial susceptibility tests using ten (10) commonly prescribed antibiotics indicated that resistance to augmentin was the highest, followed by cloxacillin, erythromycin and cefuroxime with 46 (92%), 45 (90%), 43 (86%) and 42 (84%) of the isolates respectively being resistant. However, oflaxacin and gentamicin showed great activity against the organisms as 45 (90%) of the isolates were susceptible to both antibiotics (Table 2). Graphical representation of antibiotics susceptibility is shown in Figure 1 and indicates high level of intermediate resistance to Ceftriaxone with 40 (80%) of the isolates being intermediately resistant.
Antibiotics |
Codes |
Resistant |
Intermediate |
Susceptible |
Ceftazidime |
CAZ |
39 (78.00%) |
7 (14.00%) |
4 (8.00%) |
Cefuroxime |
CRX |
41 (84.00%) |
0 (0.00%) |
8 (16.00%) |
Gentamycin |
GEN |
5 (10.00%) |
0 (0.00%) |
45 (90.00%) |
Ceftriaxone |
CTR |
5 (10.00%) |
40 (80.00%) |
5 (10.00%) |
Erythromycin |
ERY |
43 (86.00%) |
5 (10.00%) |
2 (4.00%) |
Cloxacillin |
CXC |
45 (90.00%) |
0 (0.00%) |
5 (10.00%) |
Oflaxacin |
OFL |
3 (6.00%) |
2 (4.00%) |
45 (90.00%) |
Augmentin |
AUG |
46 (92.00%) |
0 (0.00%) |
4 (8.00%) |
Novobiocin |
NOV |
30 (60.00%) |
0 (0.00%) |
20 (40.00%) |
Vancomycin |
VAN |
23 (46.00%) |
0 (0.00%) |
27 (54.00%) |
Table 2 Anti-microbial sensitivity pattern of Staphylococcus isolates
*Resistance patterns determined based on CSLI Standards.
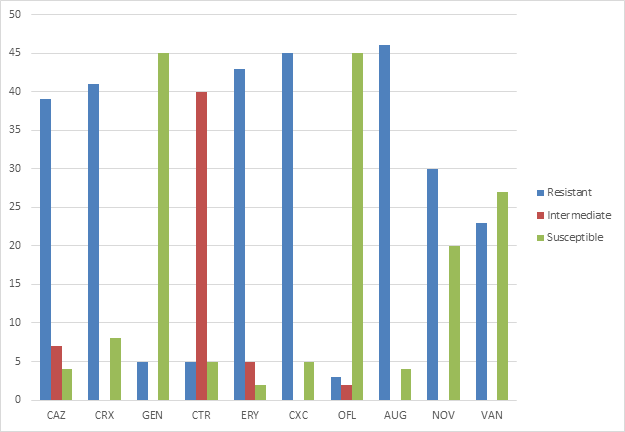
Figure 1 Antibiotics susceptibility of Staphylococci isolates.
CAZ, Ceftazidime; CRX, Cefuroxime; GEN, Gentamicin; CTR, Ceftriaxone; ERY, Erythromycin; CXC, Cloxacillin; OFL, Oflaxacin; AUG, Augmentin; NOV, Novobiocin; VAN, Vancomycin
Twenty eight (28) multidrug resistant (resistance to 2 or more classes of antibiotics) isolates were subjected to the assay for biofilm formation using the congo red agar method and 10 (35.71%) were biofilm formers indicated by the blackening of the agar while 18 (64.29%) were non biofilm formers indicated by no change in color of the agar (Figure 2). The strains displayed red colonies on medium typical for biofilm in staphylococcus isolates after 24hours of incubation. Table 3 shows the biofilm formers and the corresponding number of antibiotics to which they are resistant.

Figure 2 Screening of biofilm producers by Congo red agar medium.
|
Isolates code |
Number of antibiotics to which isolates are resistant |
Biofilm production |
|
AM10 |
7 |
-VE |
|
AM8 |
4 |
-VE |
|
AF7 |
5 |
-VE |
|
AM15 |
6 |
+VE |
|
NF3 |
6 |
-VE |
|
NM29 |
2 |
-VE |
|
NF25 |
7 |
-VE |
|
NM11 |
6 |
-VE |
|
NF20 |
5 |
+VE |
|
NM15 |
7 |
-VE |
|
NM8 |
4 |
-VE |
|
AM5 |
8 |
-VE |
|
AM3 |
7 |
+VE |
|
NM7 |
5 |
-VE |
|
NF10 |
5 |
-VE |
|
AM14 |
6 |
-VE |
|
AF9 |
7 |
+VE |
|
AM11 |
7 |
+VE |
|
NF2 |
5 |
-VE |
|
NM10 |
6 |
+VE |
|
AM9 |
7 |
+VE |
|
NF29 |
5 |
+VE |
|
AM2 |
5 |
-VE |
|
AM12 |
6 |
-VE |
|
AF12 |
5 |
-VE |
|
NM16 |
6 |
+VE |
|
AF5 |
7 |
-VE |
|
NF8 |
7 |
+VE |
Table 3 Biofilm Production by MDR Staphylococcus isolates as assayed by Congo red agar method
NM, Nasal swab from male; NF, Nasal swab from female; AM, Armpit swab from male; AF, Armpit swab from female; +VE, positive; -VE, negative
Of the six MDR staphylococci isolates assayed for biofim production, only 1(16.67%) of the isolates fluorescence indicating absence of efflux pump while the other 5 (83.33%) of the isolates did not retain the local blue dye inside the cells, thus, did not show florescence indicating efflux pump activity (Figure 3).
The existence of asymptomatic carriers is of utmost importance in the epidemiology of multidrug resistant Staphylococcus spp. These are people which are not diseased, but are colonized by multidrug resistant strains, meaning that the bacteria are part of the microbiota of the skin and mucous membranes. The nasal mucous membrane is considered the primary location colonized by multidrug resistant Staphylococcus strains.19
In this study, the carriage rate of coagulase positive Staphylococcus (CoPS) and coagulase negative Staphylococcus (CoNS) was 7.78% and 92.22% respectively among apparently asymptomatic students. The carriage rate of CoPS observed is lower than obtained in a study in Ekpoma, Edo state which reported 35.4% among apparently healthy residents of the town.20 The carriage rate is equally lower than another study in Benin city, Nigeria which reported 42.3% observed for CoPS.21 The colonization rate of CoNS was comparatively higher than CoPS in this study and approximately 12 times the rate observed for CoPS. This finding is similar to an Iranian study among students where nasal colonization rate was reported as 71.1%.22 A carriage rate of 20.6% had been reported in Benin city, Nigeria21 and 6.25% reported earlier among personnel and students in Ile-ife.23
Antimicrobial resistance has become an important public health problem in Nigeria as it has spread all over the world, limiting the use of antimicrobial drugs in the treatment of infectious diseases. However, monitoring the presence of multidrug resistant strains in certain populations may be a great step in winning the battle against high prevalence of multidrug resistant Staphylococci. The antibiotics commonly prescribed included in this study are ceftazidime, cefuroxime, gentamicin, Ceftriaxone, erythromycin, cloxacillin, oflaxacin, augmentin, novobiocin and vancomycin. Phenotypic resistance pattern of tested isolates showed a high level of resistance to multiple classes of antibiotics. Among the analyzed staphylococci isolates, none of the then were susceptible to all the tested antimicrobial agents, 1 isolate (2%) was resistant to one, 1(2%) isolate was resistant to two, 1 (2%) isolate was resistant to three, 8 (16%) isolates were resistant to four, 12 (24%) isolates were resistant to five, 12 (24%) isolates to six, 12 (24%) isolates to seven and yet 3 (6%) to eight antimicrobial agents tested in this study.
The organisms showed poor susceptibility to most antibacterial agents tested in this study. Gentamicin and oflaxacin were however the most active antibacterial agents against the organisms showing 90% susceptibility. This susceptibility profile of the tested organism to gentamicin in this study is similar to a study in Ekpoma which observed 100% susceptibility to gentamicin20 and another study which observed 75% susceptibility of methicillin resistant CoNS to gentamicin.21
It was observed in the study, that staphylococci strains showed marked resistance to all β-lactam agents tested. Resistance to augmentin, cloxacillin, cefuroxime and ceftazidime were found to be 92%, 90%, 84% and 78% respectively. Community associated CoNS and CoPS are often resistant to β-lactams.24 In a study, 80% resistance to ampicilllin in CoNS was observed, while resistance to ceftriaxone was observed in 58% of the isolates.25 Reduced susceptibility to β-lactams is usually caused by a constitutive or induced production of penicillinase, coded by blaZ. Resistance to cloxacillin is an indication/measure of methicillin resistance. Thus, indicating that the cloxacillin resistant isolates found in the nares and armpits of the asymptomatic students are either methicillin–resistant CoNS or methicillin-resistant CoPS.
Resistance to erythromycin was found to be 86% which was similar to the study of Boamah and co workers.26 Such large number of strains resistant to erythromycin may result from the fact that this is the oldest antibiotic from the macrolides that has been used in medical treatment.27
Resistance to novobiocin was high with 60% of the isolates being resistance. Only those Staphylococci belonging to the saprophyticus and sciuri group have been found to be intrinsically resistant to novobiocin.28 However, in recent findings, other CoNS have been found to also be resistant to the drug and can be linked back to mutation in the gyrase B. Single mutation confer a 32- to 64- fold decreased susceptibility to novobiocin. Since ATPase activity of mutant gyrase B subunits still has to be sufficient to support the supercoiling activity of DNA gyrase, only amino acids not directly involved in ATP binding are mutated in drug resistant isolates.29
Owing to increased resistance to methicillin among CoNS, vancomycin is frequently considered as the choice in antimicrobial therapy.30 In this study, susceptibility to vancomycin was recorded to be 46% and this differ to a study carried out in Jordan31 and Iran32 that recorded 100% susceptibility. High resistance to vancomycin was recorded in a study in Yenagoa, Nigeria to be 69.6%33 and Vancomycin resistance is still infrequent in CoNS. However, heterogeneous resistance was reported among CoNS and was associated with failure of vancomycin therapy. These heteroresistant microcolonies may be a precursor of vancomycin resistance.34
The high level of multidrug resistant Staphylococci observed in this study could be due to the misuse of antibiotics in this environment. Many Staphylococcus spp. are part of the normal bacterial flora in humans and animals.35 Several of these commensals and non pathogenic Staphylococci have been implicated in infections. According to WHO, many of such species have also been reported as multidrug resistant, which has resulted in increased cost of treating infections and increased disease burden in humans. There have been reports of increasing resistance of Staphylococcus spp. to several essential antibiotics over the past 30 years36 in different countries including Nigeria. Reasons for this resistance could range from over prescription of antibiotics, use of substandard drug (fake drugs), use of over- the- counter drugs without strict regulation and incomplete dosage on the part of the patients. These factors have thus contributed immensely to the level of MDR strains in the community. These staphylococcus species isolated from asymptomatic persons are reservoirs of antibiotics resistant genes and the time is coming when the medical world would be embattled by this emerging group of infection causing organisms.
Coagulase negative Staphylococci are also known to form biofilms and this reduces the effect of antimicrobial agents against them.36 Slime production is a great tool for antibiotics resistance among CoNS as it confers both the opportunity for transfer of resistant gene and protection against antibiotics. In the current study, 28 isolates of MDR Staphylococcus spp. were exposed to biofilm production assay and 10 (34%) were found to be biofilm producers. This value is lesser compared to findings of Martini et al.,37 who found 43.75% in clinical isolates and findings by Rathanin et al.,38 who found 90.8% biofilm producers in community isolates as tested by CRA methods. However, studies by Mathur et al.,39 and Taj et al.,40 reported a low percentage of biofilm producers using the CRA method. Those isolates that formed biofilm were found to be resistant to a large number of antibiotics (ranging from 5 to 7 antibiotics), and almost all classes of antibiotics. The results show that biofilm production is a great source of antibiotics resistance and that the susceptibility of isolates to Staphylococcus spp. is linked to biofilm production (i.e Organisms that produce biofilm show lesser sensitivity to antibiotics).41
In this study, the efflux activity among the isolates was high with 5 (83%) out of 6 isolates being positive for efflux activity using a modified form of the EtBr cartwheel test. Using a commercial blue dye instead of ethidium bromide, the efflux activity of the isolates was done. This is the first time that such modification would be done and the result obtained is of comparable standards with the non modified pattern. A study by Costa et al.,42 reported 76% of isolates were positive for efflux pump activity. Efflux pumps are of importance in antibiotics resistance because they can extrude the majority of clinically relevant antibiotics from within cells to the extracellular environment. These bacteria rely on biofilm and efflux pump activity to protect itself from different toxic material and antibiotics. Exopolysaccharides, the main component of biofilm, slow down or prevent the penetration of antibiotics to the interior cells embedded in biofilm, while the efflux pumps keep toxic materials and antibiotics under the lethal level by extruding them to the outside of the bacterial cells.43 Multidrug resistance pumps confers resistance not just to antibiotics but also to bile, hormones and other substances produced by the host and may play a role in host colonization.44
The result of this study shows that biofilm production and efflux pump are mechanisms used by Staphylococcus spp. to resist the effect of antibiotics. The antibiotic resistance of bacteria in biofilm communities has increased drastically and contributes to chronic infections. Emergence of multidrug resistance in this once thought to be commensals is of concern in the global community. The fact that these multidrug isolates exists in asymptomatic persons in the community poses a challenge for diagnosis and management. Biofilm formation in healthcare is an issue of considerable concern as it leads to mortality and morbidity. Biofilms confer ability to resist conventional antimicrobial therapies to these isolates and thus leads to persistent infections. Efflux pumps are largely conserved in bacteria for self- defense and can be a potential target for effective antimicrobial therapy to treat infectious diseases caused by multidrug resistant bacteria. Efflux activity in these organisms has increased over the years as they keep collecting the plasmid encoding some of these efflux genes from the environment. These characteristics have made these organisms reservoir of resistance genes and are often neglected as disease causative agents However, there are only few studies on the contribution of efflux to resistance phenotypes and thus lead to lack of solid knowledge of its mechanism in Staphylococcus spp.
None.
Authors declare that there is no conflict of interest.

©2020 Favour, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.