Journal of
eISSN: 2373-437X


Research Article Volume 8 Issue 6
Amity Institute of Microbial Technology, Amity University, India
Correspondence: Menaka Devi Salam, Amity Institute of Microbial Technology, Amity University, Sec 125 Noida, Uttar Pradesh, India
Received: December 01, 2020 | Published: December 22, 2020
Citation: Salam M, Varma A, Chaudhary D, et al. Novel Arsenic resistant bacterium Sporosarcina luteola M10 having potential bioremediation properties. J Microbiol Exp. 2020;8(6):213-218. DOI: 10.15406/jmen.2020.08.00311
Arsenic (As) is a highly toxic element with great mobility in the environment. If present in high concentrations in soil as well as groundwater, it poses a threat to all living organisms. Although there are many remedial methods which mostly rely on adsorption and filtration, novel technologies using microorganisms are of great attention due to their efficient degradation properties and cost-effectiveness. The present study emphasizes on novel arsenic resistant bacterium which has been isolated from electronic waste contaminated soil samples of Mandoli area in Delhi NCR, India. The isolated bacterium, identified as Sporosarcina luteola through 16S rRNA gene sequencing is tolerant to high levels of arsenic oxyanions. This bacterium designated as Sporosarcina luteola M10 could tolerate arsenate (V) upto 0.2M and arsenite (III) upto 0.01M in minimal medium. The arsenic removal efficiency was 60% of arsenate and 55.5% of arsenite respectively from arsenic amended media at 72 h as detected by atomic absorption spectroscopy. arsC, arsB and aoxB genes encoding arsenate reductase of 280 bp, arsenite transporter of 750 bp and arsenite oxidase of 450 bp respectively were found to be present through PCR amplification of genomic DNA. This is the first report of Sporosarcina luteola extremely resistant to arsenic having potential bioremediation properties.
Keywords: arsenic resistance genes, arsC gene, arsB gene, aoxB gene, arsenic removal efficiency, bioremediation
Arsenic (As) is a metalloid which is present in the environment naturally but in low concentrations within a permissible limit except in some places reporting higher levels that have led to environmental discrepancies. The presence of this element in higher concentrations can be related to enhanced contamination levels apart from natural geological conditions, industrial processes and other human activities.1 At high concentrations, it can be harmful to the flora and fauna of the ecosystem and to humans as well.2 Therefore, arsenic toxicity has become a major concern in accordance with the human health. Arsenic can exist as organic or inorganic form. The inorganic form of arsenic is generally more toxic. The predominant forms in the soil are the oxy-anions present in the oxidation states: arsenite (III) and arsenate (V). Arsenic contamination occurs in drinking water and human food, for example, fish or crops, cultivated with As-contaminated water. According to the United States Environmental Protection Agency (US EPA), it was declared in 2001 that the maximum contaminant level of As is 10μg/L in public drinking water.3 It is harmful to all life forms mainly due to its interference with many phosphate-requiring metabolic reactions, including synthesis of adenosine triphosphate (ATP). The harmful effects of arsenic to human health are cutaneous lesions (pigmentation and keratosis), carcinomas (skin, lung, bladder, liver, and kidney) and also hormonal imbalances.4,5 Various studies have been done to find out the consequences of arsenic contamination in ground water and soil on human health. In India, arsenic contamination of groundwater has been extensively reported in West Bengal. The other states which are reported to contain arsenic contamination in groundwater are Assam, Manipur, Nagaland, Tripura, Uttar Pradesh, Bihar, Jharkhand, Punjab, Haryana, Himachal Pradesh, and Rajasthan.6 The main reason is natural geological contamination such as desorption of naturally occurring minerals rich in arsenic.5
In West Bengal, arsenic concentration was 200μg/L (approximately) and was found to be associated with increased risk for stillbirth. Respiratory diseases, liver diseases, hypertension, and cancer were associated with the arsenic toxicity.7 With the rapid advancement and the immense use of electronic devices all over the globe, E-waste has become one of the fast growing types of waste and it has been reported that about 44.7 million tonnes of E-waste per year are produced worldwide in 2016. Only 20% of these wastes are properly recycled and remaining 80% are traded and processed in illegal recycling sites of developing countries.8 Heavy metals comprise about 60% of Waste in Electrical and Electronic equipment.9 They have been found to accumulate in the soils of local environments at the E-waste recycling sites. The common heavy metals detected in circuit boards of electronic devices are lead, chromium, arsenic, cadmium, mercury along with other toxic chemicals like halogenated compounds. Among them, arsenic has been reported to have high ecological risk.10 It also poses a health risk when contaminated with ground water and through the food chain, starting from aquatic organisms like small fishes and phytoplanktons which absorb the metal through contaminated groundwater, gets biomagnified.11 Studies done at an E-waste circular economy park in Tianjin, China, showed that there was a positive correlation between arsenic concentrations in the soil and dead bacteria concentrations as soil properties also change along with increase in concentrations of heavy metals i.e. as As (III) concentration increases in the soil, organic matter increases and pH of the soil decreases.12 Soil samples of illegal E-waste recycling sites of Delhi NCR, India, have also reported to contain arsenic, nickel and chromium as 3.15, 89.4 and 35.5 mgkg-1 respectively which was much higher than that of the control sample collected from a site where no E-waste activity was carried out.13
Various technologies have been developed for the removal of arsenic from contaminated groundwater, such as adsorption, precipitation, membrane filtration, bioremediation, and hybrid methods of physical, chemical, and biological techniques.14,15 Physical methods have lower removal efficiencies; on the other hand, recent advanced chemical approaches have high removal efficiencies but could be quite expensive especially for the remediation of large areas.16 Bioremediation of soils contaminated with arsenic have gained increasing interest due to its cost-effectiveness and reduction of waste. In environment there exists many types of biogeochemical cycles in which microorganisms have an important role because of their extensive and multiple metabolic activities so they can be used in bioremediation approaches because the conventional methods can only transform heavy metals from one form to other but are not able to degrade them whereas microorganisms have evolved resistance to the heavy metals during the course of evolution and exposure to heavy metals. Thus, microbial bioremediation processes are very effective and provide low-cost strategies. The approach of engineered bioremediation with optimized proliferation of bioremediating microorganisms has been beneficial in highly contaminated areas.16
Several different microorganisms have been reported to resist arsenic and also shown to remove arsenic oxyanions As (III) and As (V) from the growth medium. Pseudomas putida, C. glutamicum and Rhodococcus fascians were reported to tolerate As (III) upto 7mM, 12mM and 15mM respectively and As (V) upto 200 to 500mM.17 Many plant growth promoting rhizobacteria have also been reported to show arsenic resistance and amelioration of arsenic toxicity in plants, such as Bacillus flaxus tolerating upto 280mM As(V) and 32mMAs(III),18 Acinetobacter lwoffii (RJB-2) tolerating 125mM As(V) and 50mM As(III),19 Micrococcus luteus tolerating 275 mM As(V) and 55mM As(III).20 Among fungi, Aspergillus flavus 300 ppm As(III), Trichoderma sp. 650 ppm As(III), Rhizoglomus sp. 100ppm As(V).21
The common mechanisms for arsenic detoxification by resistant microorganisms are (a) uptake of As(V) by the transmembrane phosphate transporters like the Pit and Pst proteins, (b) uptake of As(III) by aquaglyceroporins like the glycerol facilitator GlpF homologues, (c) reduction of As(V) to As(III) by arsenate reductase ArsC, and (d) extrusion or sequestration of As(III).22 A number of different microorganisms have been isolated from arsenic contaminated environments and their mechanisms of arsenic resistance have been studied.23,24 The microorganisms also possess detoxification operons which comprise of arsC (arsenate reductase gene), arsR (transcriptional repressor), and arsB (As(III)-translocating ATPase). Some of them also contain arsD-metallochaperone and arsA-ATPase which helps in binding cytosolic As(III) and transporting it out of the cell.16 Some other microorgansims reduce arsenate to arsenite in anaerobic condition with the arsenate acting as terminal elecron acceptor.25 Two other families of arsenate resistant microorganasims are the thioredoxin (Trx) clade and Arr2p arsenate reductase. Also, ArxA (arsenite oxidase) enzymes have been reported in Alkalilimnicola ehrlichii MLHE-1 which can couple arsenite oxidation with nitrate reduction.26 Meanwhile, a number of archaebacteria and protobacteria strains have been reported to have the ability to oxidize arsenite to the less toxic arsenate. The genes encoding arsenite oxidase are aoxA and aoxB genes.16 Thus, according to the data obtained through various studies of genes involved in arsenic metabolism, the best characterized genes include those of the ars (arsenate reduction), aio (arsenite oxidation), and arr operons and are found in both plasmids and chromosomes. Microorganisms harbouring ars operon are called arsenate-resistant microbes (ARMs), those having arr operons are referred to as dissimilatory arsenate-reducing prokaryotes (DARPs) and the aio operon, formerly referred to as aro, has a system for arsenite oxidation to arsenate.27Another group which was discovered in the recent years is the oxidation system involving arx genes in which arsenite oxidation is combined with nitrate respiration.26 The arsenic resistance and metabolism genes have been found to be located in chromosomes, plasmids or both.28 In Thiomonas sp., arsenic resistance genes have been found to be present in mobile genetic elements like transposons and genomic islands.29
Developments in molecular genomics has enhanced the understanding of molecular entities and has generated vital data on the prokaryotic arsenic resistant mechanisms and the genetic bases of arsenate metabolism. Exploring the uncultured microorganisms through metagenomics has also helped in finding out the range of microorganisms present in arsenic-contaminated environments.30 This approach has validated the survival of microorganisms in arsenic contaminated environment depending on their adaptation, metabolism and interaction abilities. In the present study, an arsenic tolerant bacterium has been isolated from soil samples collected from electronic waste contaminated site of Mandoli district of Delhi NCR, India. The isolated bacterium was identified as Sporosarcina luteola through 16S rRNA gene sequencing. The tolerance of this bacterium to arsenate (V) and arsenite (III) were studied and their bioremediation capabilities were studied. Finally, the presence of important arsenic resistant genes such as arsC, arsB and aoxB were investigated.
Study site and arsenic contamination
Soil samples chosen for the study were collected from two different E-waste dumping and recycling sites in Delhi NCR where illegal recycling processes have been carried for a significant period of time. Arsenic contamination of the soil samples were already detected in our previous studies.13 Arsenic concentration of Mandoli region ranged between 1.43 to 7.93 ppm. A composite sample of 500 g was processed and sieved. The soil sample was then used for isolation of arsenic tolerant microorganisms.
Chemicals and stock solutions
Media components and chemicals used in the present study were obtained from Himedia Laboratories, Mumbai, India. Sodium arsenite (NaAsO2) was used as the source of As (III) and sodium arsenate (Na2HAsO4.7H2O) was used for As (V). Stock solutions were prepared and stored in the dark at 4°C.
Isolation of arsenic resistant bacteria
Isolation of bacteria from the soil samples was carried out using enrichment culture technique. Soil samples (10g) were added to flasks containing basal salt minimal medium (100ml) supplemented with 20mmol l-1 of As (V) or 5mmol l-1 of As (III). The composition of the basal salt minimal medium with yeast extract (BSMY)31 was as follows (in g l-1): KH2PO4 (0.05), K2HPO4 (0.05), NaCl (0.1), (NH4)2SO4 (0.3), CaCl2.2H2O (0.2), MgSO4.7H2O (0.14), yeast extract (0.1), glucose (10) and trace elements per litre as 0.6 mg H3BO3, 0.17mg CoCl2.6H2O, 0.09 mg CuCl2.2H2O, 0.1 mg MnCl2.4H2O, 0.22 mg ZnCl2. The culture flasks were incubated under shaking conditions (150 rev min-1) at 30°C for 2 days. Ten millilitre of each of these enrichment cultures were transferred into fresh medium and then incubated under the same conditions. Appropriate dilutions of enrichment cultures were plated on minimal media agar plates containing 20mmol l-1 of As(V) or 5 mmol l-1 of As(III) and the plates were incubated at 30°C for 2 days. After incubation, bacterial colonies which were morphologically dissimilar were selected from the plates and streaked onto the same medium to obtain pure cultures. Gram’s staining and biochemical tests were also done to find out the different bacterial colonies which were isolated. Isolates were maintained in glycerol stocks at -70°C.
Arsenic tolerance capacity and bioremediation test
The As tolerant capacity of the bacterial isolates were determined by growing them in 15ml minimal medium broth containing different concentrations of As (V) ranging from 10000 to 80000 mgl-1 and As (III) ranging from 500 to 2000 mgl-1 in 50ml conical flasks. The cultures were incubated at 30°C for 48h in a shaker incubator at 150 rev min-1. The growth of the cultures at different concentrations of arsenic was measured by spectrophotometric method at 600nm. The experiment was performed in triplicates. For the bioremediation test, the best isolates were selected according to their tolerance capacity and grown in minimal media containing 0.1M sodium arsenate and 0.001M sodium arsenite for 72h at 30°C under shaking conditions. The cultures were centrifuged at 10,000rpm for 10min after 24 h, 48 h and 72 h of incubation and the culture supernatant were determined for arsenic concentration by atomic absorption spectroscopy. Un inoculated minimal media containing 0.1M sodium arsenate and 0.001M sodium arsenite were also used as blank for comparision of results with the inoculated media and arsenic concentrations were measured for them by AAS.
16S rDNA sequencing of the bacterial isolate M10
From the pure culture pellet of isolate M10, genomic DNA was extracted using HiPurA™ Bacterial Genomic DNA Purification Kit (Himedia, Mumbai, India) Genomic DNA isolation kit following the manufacturer’s instructions. The Nano drop quantitative analysis of the DNA was done to check the purity of the DNA. The nearly 1.5 kb 16S-rDNA fragment was amplified using the primer set 27F (5ʹ-AGAGTTTGATCCTGGCTCAG -3ʹ) and 1492R (5ʹ-ACGGCTACCTTGTTACGACTT-3ʹ). PCR was performed in a thermocycler (Bio-Rad) with the amplification mixture containing 0.25μM each primer, 3 ng genomic DNA and 1X Dream Taq PCR Master mix (Thermo Fisher Scientific, USA) in a 40µl final reaction volume. Control PCR was set up using genomic DNA (gDNA) from E. coli BL-21DE3 and a negative control was set up with nuclease free water in place of template DNA. PCR was performed using the conditions 94°C for 3min as initial denaturation followed by 25 cycles of 94°C, 1 min), 50°C for 45 s), 72°C for 2 min and a final extension at 72°C for 10 min. The PCR product was purified by QIA quick PCR Purification Kit (QIAGEN, Inc.). Sequencing of the amplified product was carried out in the DNA Sequencing Facility at Department of Biochemistry, University of Delhi, South Campus, New Delhi, India. The 16S rDNAgene sequence was BLAST searched against GenBank database (http://www.ncbi.nlm.nih.gov/).
PCR amplification of arsenic resistance genes
The presence of different arsenic-resistance genes in genomic DNA was checked using gene specific primers for arsenite oxidase, arsenite transporter and arsenate reductase (Table 1). The PCR conditions were provided as given in Table 2.
Primer |
Sequence |
Gene name |
Amplicon lenght (bp) |
Reference |
1F |
5ʹ-GTSGGBTGYGGMTAYCABGYCTA-3ʹ |
Arsenite oxidase |
~500 bp |
32 |
1R |
5ʹ-TTGTASGCBGGNCGRTTRTGRAT-3ʹ |
|||
darsB1 F |
5ʹ-GGTGTGGAACATCGTCTGGAAYGCN AC-3ʹ |
Arsenite transporter |
~750 bp |
33 |
darsB1 R |
5ʹ-CAGGCCGTACACCACCAGRTACATN CC-3ʹ |
|||
amlt- 42-f |
5ʹ-TCGCGTAATACGCTGGAGAT-3ʹ |
Arsenate reductase |
~300 bp |
34, 35 |
amlt- 376-r |
5ʹ-ACT TTC TCG CCG TCT TCC TT-3ʹ |
|||
arsC-4F |
5ʹ-TCH TGY CGH AGY CAA ATG GCH GAA G-3ʹ |
|||
arsC-4R |
5ʹ-GCN GGA TCV TCR AAW CCC CAR TG-3ʹ |
|||
arsC-5F |
5ʹ-GGH AAY TCH TGY CGN AGY CAA ATG GC-3ʹ |
|||
arsC- 5R |
5ʹ- GCNGGATCVTCRAAWCCCCARNWC-3ʹ |
Table 1 Primers used during amplification of arsenic resistance genes
(M=A or C; N=A, C, G or T; R=A or G; V=A, C or G; Y=C or T); F, forward primer; R, reverse primer
Step |
Temperature |
Duration |
Cycles |
Initial denaturation |
95℃ |
5 min |
1 |
Denaturation |
95℃ |
90 sec |
35 |
Annealing |
*46℃ |
90 sec |
|
Extension |
72℃ |
3min |
|
Final extension |
72℃ |
5 min |
1 |
Hold |
4-10℃ |
∞ |
1 |
Table 2 PCR conditions (arsC 4f-4r)
*60°C for primers arsC 5f-5r and amlt 42f-376r, 56°C for 1F-1R, 59°C for ars B-1F, 1R
Isolation of arsenic resistant bacteria
Twelve different isolates which were morphologically different were obtained in the medium amended with arsenate. They were designated as isolates M1 to M12. They were purified by streaking on plates amended with 20 mmol l-1 of As(V).
Arsenic tolerance capacity and bioremediation test
It was found that the isolates M2, M3, M5, M7 and M10 had maximum tolerance to arsenate and arsenite. Isolate M10 was able to tolerate As (V) upto 0.2M and As (III) upto 0.01M in minimal medium. The minimum inhibitory concentration (MIC) of arsenate and arsenite against isolate M10 were 70000 mgL-1 and 1300 mgL-1 respectively (Figure 1). The culture showed decreasing growth pattern with increasing concentrations of arsenate and arsenite and finally stopped growing at 70000 mgL-1 As (V) and 1300 mgL-1 As (III). The bioremediation test results showed that isolate M10 had the maximum removal capacity of around 60% of arsenate and 55.5% of arsenite in 72 h as detected by AAS (Figure 2). The arsenate and arsenite concentrations were measured using AAS with a wavelength of 193.7 nm (0.5 nm slit) for arsenic. Isolate M10 was further studied for its genomic characterization.
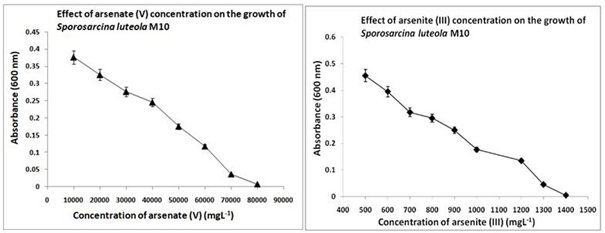
Figure 1 Plot showing MIC determination of (A) arsenate and (B) arsenite against Sporosarcina luteola M10.
16S rDNA sequencing of the bacterial isolate M10
Molecular identification of the best isolate M10 (Figure 3) carried out thorugh 16S rDNA sequencing revealed that the bacterium was Sporosarcina luteola. The nucleotide sequence of the 16S rDNA gene was submitted to Gene Bank where it appears with accession number MH084955.
Genomic DNA isolation and PCR amplification of arsenic resistance genes
By using specific primers, arsC, arsB and aoxB genes which code for arsenate reductase, arsenite transporter and arsenite oxidase respectively have been amplified (as shown in Figure 4 and Figure 5). The sizes of the amplified products were 280 bp, 750 bp and 450 bp for arsC, arsB and aoxB genes respectively. BLAST analysis of the sequences of amplified products showed 96 to 99 % similarities with the corresponding genes available in the database. The arsC gene showed maximum similarity of 98.23% with the arsenate reductase gene (Genbank Accession No.DQ517939) of Bacillus cereus strain AG24, maximum score and e-value were 494 and 0 respectively. The arsB gene showed maximum similarity of 99% with arsenite efflux pump gene (Genbank Accession No. KR051607) of uncultured bacterium clone ArsB_CfN11-9, maximum score and E value of 1325 and 0 respectively. The aoxB gene showed maximum similarity of 96.89% with arsenite oxidase genes of uncultured bacterium clone (Genbank Accession No. EF550150) and to that of arsenite oxidizing proteobacterium NT-10 (Accession No. DQ412673), Maximum score and E values were 754 and 0 respectively.
In an attempt to explore novel bacterial isolates from toxic waste contaminated sites, bacterial isolates were obtained from the E-waste contaminated sites of Delhi NCR, India. It was found that five of the isolates M2, M3, M5, M7 and M10 had maximum tolerance to arsenate and arsenite oxyanions upto 0.2M sodium arsenate and 0.01M sodium arsenite as shown by isolate M10. Also, these isolates had arsenate and arsenite removal efficiency of 40 to 60% in 72h. So far, there are meager reports on Sporosarcina luteola strains resistant to arsenic oxyanions, although one strain has been reported to be resistant to cadmium upto 5 mM. This strain of Sporosarcina luteola isolated from Wajirpur Industrial area of Delhi NCR was also reported to tolerate other heavy metals like Co, Pb, Fe and Mn.36 In a very recent study, two strains of S. luteola, UB3 and UB5 isolated from sulfide bearing tailings, were reported to sequester toxic heavy metals by bio-precipitation through the formation of metal carbonates. According to this report, UB3 and UB5 strains were found to produce metallophores including arsenophores.37 Apart from this, another species of Sporosarcina i.e. Sporosarcina ginsengisoli CR5 was reported to be tolerant to As(III) concentration of 50 mM. This bacterium exhibited mirobially induced calcite precipitation which shows its potential role in bioremediation of heavy metals.38
In the present study, out of the five highly resistant isolates, M10 which was Gram’s stain variable had the maximum as removal efficiency; therefore it was selected for further characterization of arsenic resistant genes. 16S rRNA gene sequencing results showed that the isolate is Sporosarcina luteola. The bioremediation potential and the genetic determinants for arsenic resistance were studied for the isolate designated as Sporosarcina luteola M10. Bioremediation test showed that S. luteola M10 was able to remove upto 60% of arsenate and 55.5% of arsenite from the media supplemented with sodium arsenate and sodium arsenite respectively in 72 h as detected by atomic absorption spectroscopy. The arsenic removal from the medium by Sporosarcina luteola M10 was through biotransformation and precipitation as detected by arsenic biotransforming ability and bioprecipitation tests (results not shown here). Also, it has been found that the maximum removal of arsenic was observed between pH 7 to 8. In one research study,25 Sporosarcina ginsengisoli CR5 has been studied for its arsenite removal from contaminated soil and it was found that this calcite precipitating bacterium was able to reduce As(III) to 0.88 mg kg-1 when the soil was supplemented with 500 mg kg-1. Synergistic effect of bacterial mixtures in which Sporosarcina sp. (S. soli B-22) was involved have been studied to check the removal efficiency of a mixture of heavy metals and it was found that the removal efficiency was 98% for lead and 85% for cadmium showing the increased efficiency when combined cultures are used.39 The bioreduction potential of Sporosarcina saromensis M52 for chromium, Cr(VI), was reported in which it was found to be able to remove 50-200 mg Cr(VI) in 24h. This strain was able to degrade 100 mg Cr(VI)/L completely in 24 h.40
In the present study, the genetic determinants of arsenic resistance by Sporosarcina luteola M10 were investigated using PCR amplification technique and it was found that arsC, arsB and aoxB genes which code for arsenate reductase, arsenite transporter and arsenite oxidase respectively were found to be present in the genomic DNA. The sizes of the amplified products were 280 bp, 750 bp and 450 bp respectively which correspond to the expected sizes of these genes in other bacteria.41 This results show that S. luteola M10 exhibits both arsenic reductase and oxidase activities. Also arsB gene which codes for an arsenite transporter is found to be present which is required for the bacterium’s detoxification mechanism to arsenite. The arsB and arsC genes are usually present as the ars operon in many bacteria along with arsR and arsD which are the regulatory parts.42,43 The aoxB gene which codes for arsenite oxidase is a part of arsenic metabolism in many bacteria.28 Although more studies need to be done to understand the mechanism of arsenic tolerance in S. luteola M10, this is the first study in which the arsenic resistance genes from S. luteola are being investigated.
The present study describes about a potential arsenic remediating bacterium which is extremely tolerant to arsenic oxyanions, i.e. upto 0.2 M arsenate and 0.01 M arsenite. The arsenic removal capacity of 60% arsenate and 55.5% arsenite from amended media was also quite significant showing its potential bioremediation properties. This bacterium was identified as Sporosarcina luteola and, so far, there are only few reports of Sporosarcina species which are able to remediate toxic heavy metals. The removal of arsenic from amended media shows that it has bioprecipitation or arsenic adsorption property, and further investigations are needed to prove its efficiency in arsenic removal. In the present study, the genetic determinants for arsenic resistance were investigated and three major genes of arsenic transformation have been found, namely arsenate reductase (arsC), arsenite transporter (arsB) and arsenite oxidase (aoxB). This shows that the bacterium possesses both arsenic reductase and oxidase activities in its mechanism to tolerate and survive in high arsenic concentrations. The application of this bacterium in the bioremediation of arsenic contaminated wastewaters is presently under study. Further investigations on its bioprecipitation and arsenic adsorption properties will enhance the understanding of its arsenic resistance mechanism.
We would like to acknowledge the funding provided by DST-SERB, Government of India, (File No. YSS/2015/002037/LS) for the project undertaken to study the soil microbial community and diversity in electronic waste contaminated sites.
The authors declare that there is no conflict of interest in publishing the present research work.

©2020 Salam, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.