Journal of
eISSN: 2373-437X


Research Article Volume 9 Issue 5
Biology and Horticulture Department, Bergen Community College, USA
Correspondence: Luis Jimenez, Biology and Horticulture Department, 400 Paramus Road, Paramus, New Jersey 07563, USA, Tel 2014467143
Received: September 28, 2021 | Published: October 25, 2021
Citation: Jimenez L, Peca S, Bochis J, et al. Nasal carriage of Staphylococcus aureus among a healthy suburban population: genotypic diversity and frequency of pathogenicity genes. J Microbiol Exp. 2021;9(5):159-165. DOI: 10.15406/jmen.2021.09.00337
We analyzed the frequency and genotypic diversity of Staphylococcus aureus nasal carriage in a healthy suburban population in the state of New Jersey, United States of America, from 2011 to 2018, and the presence of virulence and antibiotic resistance genes. A total of 77 isolates were analyzed by phenotypic tests and PCR testing using genes coding for S. aureus 16S rRNA, methicillin resistant (mecA), vancomycin resistant (vanA), tetracycline resistant (tetM), macrolide resistant (ermA), Panton Valentine Leukocidin (lukF), arginine catabolic element (ACME), enterotoxin A (sea), staphylococcal protein A (spa), and toxic shock syndrome (tst). Percentage of nasal carriers of S. aureus was 11% and 3% for MRSA. Based upon spa gene typing, 41 different genotypes were found. The most common types were t008, t012, and t363. Frequencies in S. aureus for spa and ACME genes were 100% and 62%. However, percentages for sea, tst, and lukF genes were 38%, 27% and 22%, respectively. The ermA and tetM were detected in 57% and 13% of isolates. None of the mecA positive isolates showed the presence of vanA. Staphylococcal cassette chromosome (SCC) mec typing was performed using a multiplex PCR. SCC mec type IV was the most common among all MRSA isolates. In conclusion, healthy individuals carried a genetically diverse population of S. aureus with different virulence and antibiotic resistance genes in the nasal cavities representing an unrecognized and understudy human reservoir for antimicrobial resistance and genotypic diversity.
Keywords: Staphylococcus aureus, nasal carriage, pathogenicity genes, antimicrobial resistance genes, PCR, genotypic diversity
Infections by Staphylococcus aureus are the number one cause for nosocomial outbreaks in the United States.1 S. aureus is responsible for causing a variety of diseases such as skin eruptions, bacteremia, endocarditis, toxic shock syndrome, and pneumonia. However, S. aureus is also part of the commensal microflora of the anterior nares in some humans.2 Most carriers are not infected by the bacteria but are reservoirs assisting the spreading of S. aureus through the community.3-5 Furthermore, nasal carriage is associated with endogenous infections and can increase the risk of nosocomial infections. Nasal carriage is influenced by a wide variety of host and bacterial factors.6 The distinction between colonization and infection is critical.7 Colonization is the presence of S. aureus without signs or illness or infection. Infection presents clinical signs of illness and inflammation. Determination of the frequency of S. aureus nasal carriage in healthy populations may lead to a better understanding of the risk factors associated with infections and the distribution of different genotypes and virulence genes. For instance, nasal carriage was shown to be an important factor in bacteremia and other diseases while elimination of S. aureus from nasal cavities reduced infection rates.
The pathogenic ability and adaptability of S. aureus to cause multiple diseases is due to the presence of a wide variety of extracellular toxins such as enterotoxins, toxic shock syndrome (TSS), hemolysins, leukocidins (Panton Valentine (PVL), and coagulase.8,9 This ability is further enhanced by the presence of the arginine catabolic element (ACME) and staphylococcal protein A (spa) genes.10,11 The ACME genes allow the dissemination of S. aureus from human skin to other parts of the body while spa genes prevent phagocytosis by white blood cells.
Antibiotic resistance is also a major factor contributing to the survival and difficulty of controlling S. aureus infections in human populations. The intrinsic resistance to antibiotics such as methicillin and other beta-lactam antibiotics is driven by the presence of the mecA gene.12 The confirmation of the presence of the mecA gene is the benchmark to diagnose methicillin-resistant S. aureus (MRSA) in hospital infections.13 The mecA gene codes for methicillin resistant by the action of a penicillin binding protein 2a (PBP2a). The mecA gene is within the mobile genetic element staphylococcal cassette chromosome mec (SCCmec) that inserts site-specifically intro the staphylococcal chromosome.
Other antibiotics such as tetracycline and macrolides are commonly used to treat S. aureus and MRSA infections.3 However, resistant strains have been isolated from clinical and environmental samples. Because of the resistance to different types of antibiotics, MRSA infections in hospital environments are difficult to treat. In some situations, the only antibiotic available for treatment is vancomycin. However, vancomycin resistant is slowly becoming a serious problem in healthcare facilities.
MRSA infections in health care environments such as hospitals, nursing homes, etc. are mostly caused by health-care associated strains (HA-MRSA).7,9 Patients with HA-MRSA infections were usually due to recent hospitalization, surgery, dialysis, indwelling medical devices, or living in nursing homes. Community-associated MRSA (CA-MRSA) infections were related to patients that did not have a history of recent hospitalization nor had any of the risk factors associated to HA-MRSA. CA-MRSA strains usually have a higher frequency of leukocidin genes and a different SCCmec element, predominantly type IV or V. Few studies have been performed to ascertain the frequency of nasal carriers of methicillin-susceptible S. aureus (MSSA) and MRSA and the distribution of virulence genes in healthy suburban populations in the United States.
The major objective of this study was to determine the numbers of nasal carriers of MSSA and MRSA and the presence of genes that enhance pathogenicity, adaptability, and survival of S. aureus in a healthy suburban population.
Study population
Seven hundred nine healthy individuals living in the suburbs of the state of New Jersey, United States of America, were included in this study that lasted from 2011 to 2018. Informed consent was obtained from all people participating in the study.
Phenotypic analysis
Nasal swab samples from one nostril were streaked on mannitol salt agar (MSA). The plates were incubated at 35°C for 48 hours. After incubation, all colonies showing mannitol fermentation (yellow colonies) were analyzed by using the Gram staining reaction, blood hemolysis on blood agar, catalase test, and the tube coagulase test. S. aureus WS obtained from Ward Scientific (www.wardsci.com) was used as a quality control strain for all tests.
DNA extractions
Chromosomal DNA extractions were performed from each isolate as previously described.14 Different aliquots of DNA were used in the PCR reactions.
Genetic identification of S. aureus isolates
Genetic identification of S. aureus isolates was performed by using 16S rRNA gene specific S. aureus DNA primers as previously described.15 The PCR reaction conditions were, an initial denaturation at 94°C for 1 min followed by 30 cycles of amplification (denaturation at 94°C for 1 min, annealing at 65°C for 3 min, and extension at 72°C for 2 min) ending with a final extension of 72°C for 5 min. A positive reaction was indicated by the presence of a 273-base pair (bp) DNA fragment.
PCR detection of pathogenicity and antibiotic resistant genes
DNA primers to detect the presence of toxic shock syndrome genes (tst) were previously described.8 The PCR reaction conditions were, an initial denaturation at 94°C for 5 min followed by 35 cycles of amplification (denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1 min) ending with a final extension of 72°C for 7 min. A positive reaction was indicated by the presence of a 326bp DNA fragment. Enterotoxin A genes (sea) were analyzed by using DNA primers previously described.8 The PCR reaction conditions were, an initial denaturation at 94°C for 5 min followed by 35 cycles of amplification (denaturation at 94°C for 2 min, annealing at 57°C for 2 min, and extension at 72°C for 1 min) ending with a final extension of 72°C for 7 min. A positive reaction was indicated by the presence of a 102bp DNA fragment. Arginine catabolic mobile element (ACME-encoded) arcA gene was analyzed by using specific primers previously described.10 The PCR reaction conditions were, an initial denaturation at 95°C for 5 min followed by 30 cycles of amplification (denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min) ending with a final extension of 72°C for 5 min. A positive reaction was indicated by the presence of a 624bp DNA fragment.
Staphylococcal Protein A (spa), mecA, and leucocidin (lukF) genes were amplified using DNA primers as previously described.12 The PCR reaction conditions were, an initial denaturation at 94°C for 5 min followed by 30 cycles of amplification (denaturation at 94°C for 30 sec, annealing at 59°C for 1 min, and extension at 72° C for 1 min) ending with a final extension of 72° C for 10 min. Positive reactions for lukF, and mecA genes were indicated by the presence of DNA fragments of 85bp and 162bp, respectively. Spa gene detection was indicated by DNA fragments ranging from 180 to 600bp. Vancomycin resistant genes (vanA) were analyzed by using DNA primers described by Azimian et al.16 The PCR reaction conditions were, an initial denaturation at 94°C for 5 min followed by 40 cycles of amplification (denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72° C for 2 min) ending with a final extension of 72°C for 5 min. A positive reaction was indicated by the presence of a 713bp DNA fragment. Tetracycline, tetM, and macrolide resistant, ermA, genes were analyzed using primers described by Strommenger et al.17 The reaction conditions were 94°C for 3 min followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, with a final extension of 72°C for 7 min. Positive reactions were indicated by the amplification of 158bp and 190bp fragments for tetM and ermA, respectively.
DNA sequencing
Sequencing of the amplified PCR fragments from bacterial isolates were performed by Genewiz, LLC (South Plainfield, New Jersey). Homology searches were performed using the GenBank server of the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the BLAST algorithm.18
Spa typing
The polymorphic X region of the spa gene was amplified from all S. aureus isolates as described above. All sequencing reactions were performed by Genewiz LLC (South Plainfield, New Jersey). Homology searches were performed using the GenBank server and the BLAST algorithm. Spa types were assigned by using the Bionumerics software application version 7.6.3 (Applied Maths Inc., Austin, TX), (Applied Maths NV Sint-Martens-Latem, Belgium) installed in a windows computer.19
SCCmec typing
Typing of the SCCmec genetic element was performed using a multiplex PCR assay described by Boyle et al.20 Amplification conditions comprised 4 min at 94°C, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, with a final extension of 4 min at 72°C. The SCCmec type was determined on the basis on the band pattern previously described.20
Characterization of S. aureus isolates by phenotypic analysis
A total of 77 isolates were described and identified as S. aureus from nasal cavities of 709 healthy individuals. All isolates and the control strain were gram positive cocci, fermented mannitol, produced coagulase, and showed beta-hemolytic reactions on blood agar.
Identification of S. aureus by 16S rRNA analysis
Genetic analysis using 16S rRNA primers confirmed the presence of the specific 273bp S. aureus 16S rRNA fragment (Figure 1). All 77 isolates and the control strain showed a positive reaction with no other DNA fragments detected. After genetic identification was completed, the nasal carriage for S. aureus was calculated to be 11%.
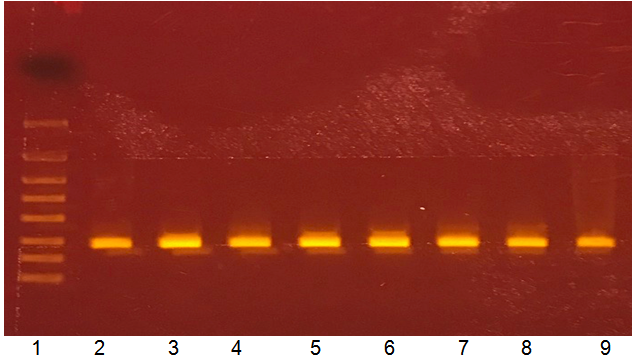
Figure 1 Genetic identification of human S. aureus nasal isolates using 16S rRNA sequences
Lanes:
Lane 1: Molecular weight marker, bp (from top to bottom, 4000, 2000, 1250, 800, 500, 300, 200, 100)
Lane 2: S. aureus 57
Lane 3: S. aureus 58
Lane 4: S. aureus 59
Lane 5: S. aureus 60
Lane 6: S. aureus 61
Lane 7: S. aureus 62
Lane 8: S. aureus 63
Lane 9: S. aureus 64.
Detection of antibiotic resistance genes
The presence of the mecA gene, indicated that 23% of the isolates were MRSA (Figure 2a). MRSA carriage was identified in 3% of the people tested. Seventy seven percent of the isolates did not carry the mecA gene. None of the mecA isolates showed a positive reaction for the presence of the vanA gene. The ermA and tetM genes were found in 57% and 13% of the S. aureus isolates. When comparing MRSA and MSSA, higher frequency of tetM genes were found in MSSA while ermA was found to be more predominant in MRSA strains (Figure 2b).

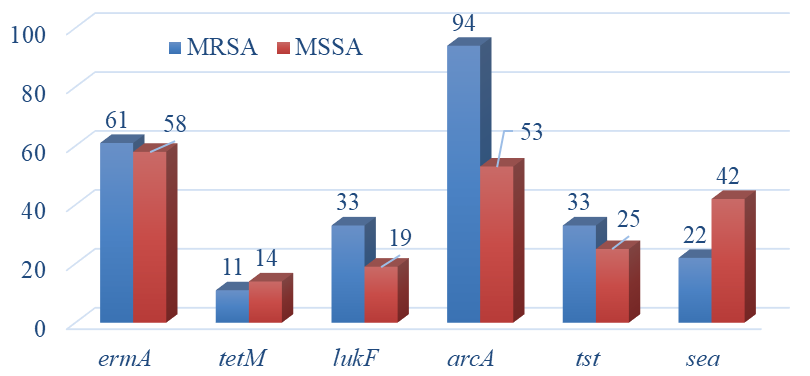
Figure 2 Frequency of pathogenicity genes.
a. Percent of positive in S. aureus population. N=77.
b. Percent of positive in MRSA (n=18) and MSSA negative (n=59)
population.
Characterization of S. aureus isolates by molecular typing techniques
SCCmec typing of MRSA isolates demonstrated that 83% contained a type IV cassette on the chromosome (Table 1). There were other two SCCmec types detected. Those were type I (6%) and III (11%).
|
Isolate |
16S rRNA |
SSCmec Type |
spa Type |
lukF |
ACME |
tst |
sea |
tetM |
ermA |
|
Sa17 |
+ |
IV |
t304 |
- |
+ |
- |
- |
- |
+ |
|
Sa25 |
+ |
IV |
t3380 |
- |
+ |
+ |
+ |
+ |
- |
|
Sa31 |
+ |
IV |
t316 |
- |
+ |
- |
- |
- |
- |
|
Sa33 |
+ |
IV |
t216 |
- |
+ |
- |
- |
- |
+ |
|
Sa35 |
+ |
I |
t4951 |
+ |
+ |
- |
- |
- |
+ |
|
Sa37 |
+ |
IV |
t443 |
+ |
+ |
- |
- |
- |
+ |
|
Sa38 |
+ |
IV |
t062 |
+ |
+ |
+ |
- |
- |
- |
|
Sa41 |
+ |
IV |
t443 |
- |
+ |
- |
- |
- |
- |
|
Sa43 |
+ |
IV |
t216 |
- |
+ |
+ |
- |
- |
- |
|
Sa45 |
+ |
III |
U* |
- |
- |
+ |
- |
- |
- |
|
Sa48 |
+ |
IV |
t008 |
+ |
+ |
- |
- |
- |
- |
|
Sa50 |
+ |
IV |
t334 |
- |
+ |
+ |
+ |
- |
+ |
|
Sa51 |
+ |
IV |
t051 |
- |
+ |
- |
+ |
- |
+ |
|
Sa53 |
+ |
IV |
U* |
+ |
+ |
- |
+ |
+ |
+ |
|
Sa55 |
+ |
IV |
t240 |
- |
+ |
- |
- |
- |
+ |
|
Sa56 |
+ |
IV |
U* |
- |
+ |
- |
- |
- |
+ |
|
Sa76 |
+ |
III |
t359 |
+ |
+ |
+ |
+ |
- |
+ |
|
Sa77 |
+ |
IV |
t1414 |
- |
+ |
- |
- |
- |
+ |
Table 1 16S rRNA, SSCmec, and spa typing with pathogenicity gene results for MRSA isolates
U*=Unknown
All 77 isolates showed the presence of the spa gene (Figure 2a). Typing of the spa gene yielded 41 different genotypes (Table 2). The most common genotypes were t008 (n=4), t012 (n=4), t021 (n=3), t338 (n=3), t363 (n=4), and t443 (n=3). All other genotypes had 1 or 2 strains. There were 15 isolates that did not belong to any spa type. The most common genotype among MRSA strains was t443 (Sa37, Sa41). Two genotypes t012 (Sa40, Sa44, Sa67, Sa72) and t363 (Sa2, Sa16, Sa22, Sa58) were the most common with MSSA strains.
|
Most frequent spa-types |
t008, t012, t363, (all n=4) |
|
t021, t338, t443, (all n=3) |
|
|
Other spa-types |
t002, t024, t209, t216, t316, t334, t571 (all n=2) |
|
t018, t051, t062, t065, t084, t160, t177, t240, t267, t304, t359, t360, t548, t688, t693, t789, t1046, t1178, t1414, t1631, t2279, t2339, t3380, t4359, t4460, t4951, t11321, t13691 (all n=1) |
|
|
|
Spa-types not in database (n=15) |
Table 2 List of spa-types and number of isolates (n)
Detection of virulence genes in S. aureus isolates
Sixty two percent of the S. aureus isolates from nasal samples showed the presence of the ACME gene (Figure 2a). Sea genes showed a frequency of 38% while tst and lukF genes showed 27% and 22%, respectively (Figure 2a). Only 27% of the MRSA strains with a type IV SCCmec cassette showed the presence of lukF genes. The ACME gene was found in all SCCmec type I and IV strains. However, it was not detected in one of the type III isolates (Table 1). Twenty nine percent of SCCmec type IV strains showed the presence of tst and sea genes.
Genes for colonization (ACME), toxic shock syndrome (tst), and leucocidin production (lukF) were found to have higher frequency in MRSA than in MSSA (Figure 2b). Only sea genes showed higher percentages with MSSA than with MRSA while spa gene percentages were similar for MRSA and MSSA. When analyzing the numbers of antimicrobial resistant and virulence genes, 39% of MRSA isolates showed the presence of at least 4 of those genes while 22% of those isolates showed 5 genes. When compared to MRSA, 17% of MSSA isolates showed the presence of 4 genes and 10% with 5 genes.
Nasal carriage of S. aureus may represent a significant risk for invasive infection in susceptible populations. The people sampled in this study were healthy with no signs or symptoms of skin, respiratory, sinus, or throat infections. S. aureus nasal carriage in the present study was found to be 11%. Previous studies in the USA showed a 35% carriage rate but that study was done by sampling both nostrils2. However, in this study we only sampled one nostril which might have underestimated the numbers of carriers. Higher percentages of S. aureus carriers were reported in Iran, 20.8%,4 China, 15.4%,3 India, 12%,21 Ghana, 22.1%,22 and Brazil, 31.1%.23 In general, the colonization rate of S. aureus in healthy individuals through the world ranges from 3 to 70%.24 However, several factors can have predisposed nasal carriage. Age, gender, health, and chronic diseases are among the factors considered to increase nasal carriage. The environment can also be a reservoir for S. aureus. Environmental contamination with S. aureus was reported where MSSA were recovered from environmental surfaces.25 In that study 28% of the people sampled were found to be S. aureus carriers.
Previous studies reported MRSA carriage values very similar to the values found in this study (3%). Kildow et al.,2 reported a 3.2% carriage rate when samples from both nostrils were analyzed. Similar percentages of MRSA carriers were found in China.3 Higher percentages of MRSA carriers were reported in India21 and New Zealand (5%).26 A U.S. population survey reported the rate of colonization of S. aureus and MRSA in the USA ranged from 31.6% to 0.84%.1 Evidently S. aureus and MRSA carriers varied significantly even within similar populations in the same countries. A major limitation of our study was the sampling of only one nostril which might have underestimated the carriage rate for MRSA.
SCCmec typing results for MRSA isolates indicated the predominant presence of a type IV cassette. SCCmec is a genetic element involved in the horizontal transfer of resistant genes that has been used as a marker for distinguishing between HA-MRSA and CA-MRSA.13 Type IV is mostly associated to CA-MRSA strains.13,27 HA-MRSA are commonly associated to types I-III. In our study 83% of MRSA strains belonged to type IV SCCmec while healthy carriers in Mexico were found to have a lower percentage of type IV strains, 21.4%.28 Studies of clinical isolates from community health centers in the New York (NY) city area showed that most infections were related to CA-MRSA containing type IV SCCmec elements.29 Most isolates belonged to the USA300 clone (t008, SCCmecIV, PVL+, AMCE+) or to closely related clones showing different spa and SCCmec types without the presence of PVL or ACME genes. Strain USA300 is the predominant MRSA clone in North America. Nasal MRSA isolates showed 84% of PVL. In this study only 1 isolate, Sa48, showed the typical USA300 clone profile. However, 3 MSSA isolates, Sa6, Sa7, and Sa18 were found to belong to the t008 spa-type but showed a negative reaction for mecA, ACME and PVL. Environmental contamination of surfaces in households in NY city and nursing homes in Ohio were reported to be major reservoirs for USA300 dissemination, infection, and diversification.30,31 However, we did not carry any environmental sampling during this study. A year after our study was completed, surfaces were sampled from different locations but no S. aureus or MRSA were isolated.
None of the MRSA isolates showed the presence of the vanA gene indicating their possible sensitivity to the antibiotic vancomycin. Vancomycin continues to be the most common antibiotic for MRSA infections when other antibiotics are inefective. However, genes coding for macrolide and tetracycline resistance were detected. Previous studies showed higher percentages of tetM genes in MRSA isolates from European clinical samples.17 Resistant to the tetracycline by the tetM gene is mediated by a non-covalent modification of the ribosomes during protein synthesis. The frequency of tetM genes in MRSA isolates from those samples was 29% while in this study we found lower values, 11%. MRSA isolates from burn unit patients in Iran also showed higher percentages of tetM genes, 32.4%.32 However, when it came to macrolide resistance genes higher percentages were found in this study, 61%, than previously reported for clinical isolates, 57%.17 Other studies in France showed slightly higher frequencies in clinical isolates of S. aureus, 63.2%.33 Only 57% of the S. aureus isolates in this study showed the presence of the ermA gene. The ermA gene codes for methylases that add methyl groups to the adenine residue at position 2085 in 23S rRNA, resulting in the significant reduction in the bonding between ribosomes and macrolide antibiotics. Clinical isolates from China showed 21.6% of MRSA strains containing ermA genes while MSSA showed a lower frequency, 11.1%.34 The same study found that MRSA isolates showed higher frequencies of tetM, 67.1%, than MSSA, 59.3%.
PVL genes are strongly associated with skin infections, soft tissue infections, and severe necrotizing pneumonia. They destroy white blood cells such as neutrophils and macrophages and promotes tissue necrosis. In this study, PVL genes were found in 22% of S. aureus isolates which was higher than studies from Indonesia (10%)35 and Iran (20%).4 However, higher prevalence of PVL genes were reported in New Zealand with 31%.26 They found similar percentages of PVL genes in clinical and nasal isolates. S. aureus isolates in Ghana were found to have much higher PVL prevalence, 58%.22 Studies in a New York prison found 93% isolates carrying PVL genes.36 Lower percentages of PVL genes were found in Iowa and Nebraska populations with only 8% of S. aureus carrying PVL genes.2 MRSA isolates in this study showed higher frequency of PVL genes, 33%, when compared to MSSA (19%). Studies in Indonesia reported a high frequency of PVL genes, 10.6%, in MSSA isolates.35 However, nasal MSSA isolates from community health centers in the NY city area showed a PVL frequency of 30%.29
Higher frequency of PVL genes is a common marker for CA-MRSA than HA-MRSA which usually showed less than 4%.9,13,37 Studies in China and Australia reported low percentages of PVL genes in CA-MRSA, 11% and 0% respectively.3,38 Studies in Tennessee found a 93.6% PVL positives among CA-MRSA.27 Shukla et al.9 demonstrated that the prevalence of PVL genes in CA-MRSA from the states of Minnesota and Wisconsin was 100%. The healthy population we studied showed CA-MRSA strains with a very low PVL frequency, 27%. Higher numbers were found in CA-MRSA isolates from a healthy Mexican population,28 67%.
Regarding the ACME gene, all type IV SCCmec strains in this study were positive with MRSA strains showing an overall frequency of 94%. Percentage of ACME was also very high for MSSA strains with 53% showing a positive reaction. ACME was not highly prevalent in a Mexican population colonized by CA-MRSA.28 S. aureus isolates from healthy Iranian populations showed lower numbers of ACME genes, 17%. The ACME gene (8.37%) was detected in MRSA strains from England and Wales isolated from skin and soft tissue infections.39 Out of 104 S. aureus isolated from wounds and nasal samples in Community Health Centers in the NY City metropolitan area, ACME was present in all USA300 strains.29
The presence of tst and sea genes can enhance the virulence of nasal S. aureus isolates. TSS is a condition characterized by fever, rash, and hypotension. In this study, MRSA isolates showed higher frequency of tst genes than MSSA. Previous studies reported a frequency of 24.3% from healthy carriers.8 Clinical S. aureus strains in Germany showed lower frequency of tst genes with 20.3%.40 We found a higher percentage, 27%, among the S. aureus population analyzed in this study. However, Nagao et al.41 reported a much higher percentage in Japanese clinical isolates with 75% of MRSA possessing the tst gene compared to 33% of the non-clinical MRSA isolates found in our study. A healthy student population in central Iran showed type IV SCCmec strains with a 28.5% prevalence.42 In this study, we found very similar abundance, 29%, in type IV SCCmec strains.
Staphylococcal enterotoxins such as sea are the main cause of food poisoning by S. aureus causing intensive intestinal peristalsis. The sea numbers, 38%, were much higher than the ones reported by previous analysis of healthy S. aureus nasal carriers.8 Analysis of Canadian and Dutch S. aureus isolates reported that only 19.6% were positive for sea. Clinical S. aureus strains in Germany showed lower percentages with only 15.9% of isolates from nasal and blood samples showing the presence of the sea gene.40 In our study, sea was the only gene along with tetM found at higher frequencies in MSSA than in MRSA. However, studies of clinical isolates from China showed MRSA with higher numbers of sea than MSSA.34
In summary, S. aureus and MRSA isolated from a healthy suburban population exhibited a very high genotypic diversity based upon typing of the spa gene. However, most MRSA strains detected belonged to SCCmec type IV. Our data provide further evidence of the frequency of antimicrobial resistance and virulence genes indicating the distribution of potentially pathogenic strains among non-symptomatic and healthy suburban populations which may represent a potential reservoir to disseminate these strains among susceptible populations. A possible limitation of our study was the sampling of only one nostril which may have underestimated the frequency of potential S. aureus and MRSA carriers.
None.
Authors declare that there is no conflict of interest.

©2021 Jimenez, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.