Journal of
eISSN: 2373-437X


Research Article Volume 7 Issue 3
Research Center of Molecular Biology, Baqiyatallah University of Medical Science, Iran
Correspondence: Dr. Ali Karami Ph.D, Research Center of Molecular Biology, Baqiyatallah University of Medical Science. P.O. Box 19945-581, Tehran-IRAN, Tel 0098-21-88039883
Received: April 24, 2019 | Published: May 30, 2019
Citation: Karami A, Pourali F. Molecular detection and identification of fatal infectious agent, isolated from patients in an outbreak occurred in Sistan & Balochistan province of Iran. J Microbiol Exp. 2019;7(3):144-148. DOI: 10.15406/jmen.2019.07.00253
An outbreak of tularemia or plague like syndrome occurred in the province of Sistan and Balochistan of Iran, from May to June 2007. Tularemia and plague had not been reported in this region for last 100 years and before. Thirteen eight cases were identified with ulcer glandular syndrome dominant to all cases with age from children to elderly ages. With fatality rate of 26% and death of 8 patient that have been reported by villagers very late, all other patients and new cases recovered after antibiotic therapy and other heath measures to prevent the spread of diseases. Targeted chemoprophylaxis, sanitation, and vector control played a crucial role in controlling the outbreak. Coco bacillus like agents was isolated from the blood samples of the patients. Epidemiologic, microbiological and molecular analysis of samples findings suggested the possible existence of a local animal reservoir, food or water contamination during this period, but its origin could not be determined. This sudden and unexpected reemergence of tularemia or plague like disease in this province with no background history of rodents or other animal death from the disease or any human cases is important for molecular epidemiology and root finding.
Tularemia is a highly infectious disease which can be caused by a small amount of bacteria; it is naturally occurring. Typically found in rural areas, it is common in animal populations, and functions as a zoonosis if transmitted via the bite of an infected animal (in particular rodents, rabbits, and hares), or if transmitted via a tick or deer fly bite.1 The disease can be contracted through the handling inhaling the bacteria which causes the illness. If inhaled and left untreated, it can lead to severe respiratory illness, pneumonia, and systemic infection. Tularemia, a zoonotic disease caused by the highly infective, virulent, non-sporulating Gram-negative coccobacillus Francisella tularensis It is a very small bacterium, 0.7-1.0 microns in length, that is non-motile and does not form spores... the organism can be isolated from contaminated environmental sources such as water and mud.2 Humans can become infected with tularemia through many different routes, and age and sex factors do not seem to affect susceptibility. Common routes of infection are: through insect bites including ticks and mosquitoes, handling of contaminated animal tissue or fluid, recurrent contact act with contaminated water, food or soil, halation of infective aerosols.
Tularemia has a 3-5 day incubation period and the initial symptoms are flu-like in nature. These symptoms include: sudden fever, chills, headache, diarrhea muscle aches, dry cough, and progressive weakness. Other symptoms are more specific and depend on the route of infection. Skin ulcers and swollen glands may occur through coetaneous exposure to tularemia. Pneumonic tularemia occurs when the bacteria is inhaled. Symptoms for this form of tularemia include chest pains, bloody sputum and difficulty breathing.2 Francisella tularensis infection occurs in Iran is supported by a study published in 1973, that serological evidence of infection was found in animals scattered throughout.3 There is no data on epidemiologic or presence of tularemia cases in sistan and baluchestan provinces.
Plague is primarily a bacterial zoonosis affecting rodents. It is caused by Yersinia pestis and is transmitted from animal to animal by fleas. Humans usually become infected through the bite of an infected rodent flea, inhalation of infected aerosols or ingestion of contaminated food and water. Bubonic plague, a severe infectious disease which, in the absence of appropriate antimicrobial drug therapy, can evolve to a rapidly fatal septicemia or pneumonia, can develop. A pneumonia form, which enables direct transmission to contacts, can be responsible for highly lethal outbreaks.4 Currently, plague natural foci persist in Asia, the Americas, and Africa (where most human cases occur).5 Plague foci have previously existed in the Kurdistan province of Iran.6
The surveillance of territories surrounding the plague focus of Kurdistan province in Iran, by inspection of wild rodent burrows, permit to reveal the existence of an epizootic in a new focus located in the Eastern Azerbaijan province, where plague was never reported. In one study 14 strain of Y. pestis were isolated from Different rodents and from the fleas. The eventual relationship between these two areas separated by about 200km was investigated: neither attempts to isolate Y. pestis nor serological surveys permit to reveal any sign of plague enzootic in the zone between the Kurdistan focus nor the new described focus of the Eastern Azerbaijan.7-10 There is no report of plague in Sistan and baluchestan of Iran.
Sistan-Balochistan province is located in extreme southeastern Iran. The combined Sistan and Baluchestan province today accounts for one of the driest regions of Iran. During May and Jun 2007, several patients with signs of severe infection and painful inflammatory adenopathy were admitted to the health branch of. Khash, saravan and other local health care centers. After eliminating all other possible differential diagnoses, clinicians suspected tularemia. Blood samples collected from patients were sent to the Microbiology Department of Zahedan tropical disease laboratory and Institute Pasteur of Iran. All samples were examined with standard bacteriologic methods. Direct examination of smears was performed after Wayson and Gram staining. Blood samples were cultured in Castaneda medium and examined daily. Further subcultures and biochemical tests were performed according to standard microbiological protocols.
PCR analysis
PCR was performed on DNA extracted from isolated bacteria from blood cultures. We have uses primers specific for F1 gene of the yersinia pestis as bellow protocols:
|
DNA template |
1ml |
|
Primer F |
1ml |
|
Primer R |
1ml |
|
DNTPs |
2ml |
|
MgCl2 |
0.75 |
|
PCR buffer 10X |
2.5 |
|
Taq DNA pol |
0.5 |
|
D.D.W |
upto 25 |
PCR program
|
Stage |
temp.ºC |
time(min) |
cycle no. |
|
Hot start |
94 |
5 |
35 |
|
Denat. |
94 |
1 |
|
|
Annea. |
62 |
1 |
|
|
Exten. |
72 |
1 |
|
|
Final Ext |
72 |
7 |
Nested PCR
We have used inner primers specific for the F1 gene sequence to confirm the PCR product. The protocol is the same as above (Table 1).
|
Biovar |
Glycerol |
Arabinose |
Nitrate |
PCR and nested PCR |
|
Antique |
+ |
+ |
+ |
+ |
|
Mediavalis + |
+ |
- |
+ |
|
|
Orientalis - |
+ |
+ |
+ |
|
|
Microtus |
+ |
- |
- |
+ |
|
Zahedan-khash |
+ |
- |
+ |
|
Table1 Biochemical characteristic of different biovars of Yersinia pestis, Iranian type is identified as Mediavalis
For final confirmation, PCR product was sequences and nucleotide sequence were analyzed by Bioinformatics programs like BLAST to compare the sequence with databases (Table 2).
|
Sequences producing significant alignments |
(bits) |
Value |
|
gi ׀ 39754771 ׀ gb ׀ AY450847.1 ׀ Yersinia pestis strain 482 plasm_ _ _ |
48 |
4e-04 |
|
gi ׀ 39754755 ׀ gb ׀ AY450846 .1 ׀ Yersinia pestis strain 1351 piss_ _ _ |
48 |
4e-04 |
|
gi ׀ 39754751 ׀ gb ׀ AY450845 .1 ׀ Yersinia pestis strain EV plasmi_ _ _ |
48 |
4e-04 |
|
gi ׀ 3883003 ׀ gb ׀ AF 074611 _ 1 ׀ Yersinia pestis is KM plasmid pliT -1 _ _ _ |
48 |
4e-04 |
|
gi ׀ 2996286 ׀ gb ׀ AF 053947 _ 1 ׀ Yersinia pestis is KM plasmid OTT 1, _ _ _ |
48 |
4e-04 |
|
gi ׀ 45357241 ׀ gb ׀ AE017045 _1 ׀ Yersinia pestis biovar Mediaeval _ _ _ |
48 |
4e-04 |
|
gi ׀ 5834685 ׀ am:do ׀ AL117211 _1 ׀ YP PliT1 Yersinia pestis C092 plasm_ _ _ |
48 |
4e-04 |
|
gi ׀ 52537981 ׀ emb ׀ AJ 698720_ 1 ׀ Yersinia pestis pG8786 plasmid |
48 |
4e-04 |
|
gi ׀ 48620 ׀ emb ׀ X61996.1 ׀ YPCAF Y. pe se is genes call, caf1M, caf _ _ _ |
48 |
4e-04 |
Table 2 Analysis of primer used in this study
Samples taken from members of one family in the same village, reveled several Gram negative coocobacill shape bacteria similar to Francisella tularensis and Yersinia pestis. Further bacteriological analysis In Institute Pasteur in Teheran demonstrated that bacteria are more similar to Yersinia pestis with Negative in Nitrate reduction and positive in Arabinose. Diagnosis of plague was suspected and confirmed by isolation of a bacterial DNA and molecular analysis. Figure 1 demonstrates the PCR product of 530 bp from F1 gene of the Yersinia pestis. Figure 2 is result of Nested PCR with inner primers to f1 gene and as its shown 2 fragment of 530 bp original gene and 280 bp is shown. The sequencing and PCR data demonstrate that Bacteria had F1 gene sequence (Figure 3-5).
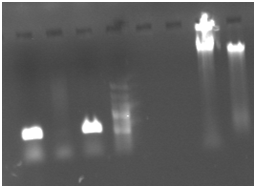
Figure 1 PCR of DNA extracted from bacteria isolated from Blood culture of patients . Primers F1 and R1 from gene specific to Yersinia pestis.
5 and 6. Total DNA extracted from bacterial samples.
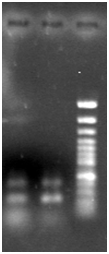
Figure 2 Nested PCR of the product from first PCR with inner primers.
Epidemiologic investigation did not identify any other tularemia or plague patients before this outbreak. It is unlikely that other cases occurred and remained undetected during this period. It seems that bacterial agent gives severe infection with high fatality rates. In this investigation we used molecular methods to diagnose identify the infectious agent of unknown origin for the first time. This coordinated experience to rapidly diagnose and contain the severe fatal infection by using molecular methods can be uses for any other similar situations. The bacteriologic diagnosis is a long procedure (at least 4 days) and, in this epidemic molecular method contributed to the effectiveness of the response. The outbreak occurred in a poor rural area of Khash and Saravan villages, with inadequate sanitation. The resident's interview revealed increase in the population of commensal rodents but no unusual rodent mortality was noted during the weeks preceding the outbreak.
A crisis management program was developed based on standardized case management, prophylactic treatment and follow-up of contacts sharing the patients, and vector control. No natural focus of tularemia or plague had ever been described in these provinces. Assumption for the original sources of infection have been claimed as contaminated meat, possible source of rabbit or other hunted animals or transported goat from border so the reappearance of human cases in this area can be explained in 3 ways: a recent importation of infected animals, contaminated meat from hunting or a sudden manifestation of a natural focus that had remained silent for decades and finally possible bioterrorist act. These has been observation and claims by villagers and officials of the region of unknown disease and death of goats in the villages central to the epidemic and other rural areas. Preliminary examination of few carcasses fund in the region by Veterinary member of the group did not ravel any evidence but there is no scientific report of tests or laboratory analysis of samples from these carcasses. So, it is not possible to exclude any of this assumption without further molecular epidemiology of the region and exact examination of samples from animals, rodents, vectors, water and environmental samples and follow up of rural population by serology tests. The outbreak occurred at the region with no cultivation or agricultural activity. The current challenge in terms of public health is to determine the source, genotype and the type exposure of people to the infectious agent.
We are grateful to Dr. Izadi President of the University Of Medical Science Of Zahedan and his team members for their support and kindness, and other members of the epidemiology team for their assistance.
Author declares that there is no conflict of interest.

©2019 Karami, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.