Journal of
eISSN: 2373-437X


Research Article Volume 4 Issue 6
1Department of Pharmacy, ASA University Bangladesh, Bangladesh
2Department of Biology, Medgar Evers College of the City University of New York, USA
Correspondence: Alam Nur E Kamal, Department of Biology, Medgar Evers College of the City University of New York, 1638 Bedford Avenue, Brooklyn, New York 11225, USA , Tel 718-270-5078
Received: April 26, 2017 | Published: May 16, 2017
Citation: Begum K, Mannan SJ, Rahman MM, Mitchel-Antoine A, Opoku R, et al. (2017) Identification of Antibiotic Producing Bacteria from Soil Samples of Dhaka, Bangladesh. J Microbiol Exp 4(6): 00134. DOI: 10.15406/jmen.2017.04.00134
Multidrug-resistance in bacteria is a serious and common problem that demands an urgent need to discover new antibiotics. In recent years multiple approaches have been taken to develop novel, potent, and less toxic antibiotics from natural sources against pathogenic bacteria. Soil has been considered as a natural source of obtaining bacteria with the ability to produce noble antibiotics. We studied soil samples in Dhaka municipal area to identify bacteria having antibiotic producing ability. We have isolated three bacterial strains (named K1, K2, and K19) producing growth inhibitory components effective against both gram positive and gram negative bacteria. Bacteria were found to produce circular, regular, white colonies. Gram staining indicated that K1, K2 was gram negative, while K19 was gram positive. Cellular morphology was studied by scanned electron microscopy (SEM) and found that K1, K2, and K19 are short rod shaped. It was found that the antibacterial factors from the bacterium are released in to the cell culture medium. The antibacterial activity of the component(s) remains active when the cell culture supernatant was heated at 45oC for five minutes. Further characterization of the bacteria and their antibacterial compounds are in progress in our laboratory.
Keywords: Soil; Bacteria; Isolation; Antibacterial compounds
TSA, trypticase soy agar; TSB, trypticase soy broth; E. coli, escherichia coli; S. aureus, staphylococcus aureus
Microbial population in soil depends of various factors such as temperature, salt concentration, pH, carbon sources etc. While growing in soil, microorganisms compete with each other for growth advantages and evolve a mechanism to win over other organisms.1 One of those mechanisms includes production of components that inhibit the growth of other microorganisms. Diversity in environment in the biosphere favor amplification of specific types of microorganism. The microbial pool therefore evolves a wide range of biochemical mechanisms which can be manipulated and utilized for the benefit of other organisms including human. Identification of such microorganisms raises a possibility of finding noble antibiotics with a potential clinical use.2,3
Bacterial pathogens (e.g. Mycobacterium tuberculosis, Staphylococcus aureus etc.) have been reported to frequently acquire resistance to antibiotics, and exhibit multi-drug resistance. In recent years multidrug-resistance in bacteria has raised a serious concern among scientific community. It highlights a greater need to find more antibiotics as well as alternative antimicrobial substances that can be used for use in clinics, food preservation and dairy products.4,5 Consequently, research on antimicrobial compounds as a new class of drugs has increased in the recent past as they exhibit both narrow and broad spectrum inhibitory activities against gram-positive and gram-negative bacteria. An attractive alternative method to discover antibacterial agents is the use of promoter-inducible reporter assays.6–8 The selective induction of the reporter fusion indicates that a compound is perturbing the pathway of interest.8 Antibiotic resistance is a great challenge to modern medicine and demands discovery of novel antibiotics.9–11 Microorganisms were found to produce secondary metabolites with a diverse chemical structure and antimicrobial activities.12,13 These findings raise a high possibility to identify bacterial strains from soil samples that might produce reagents with biochemical and pharmacological importance.
The ability of Actinomycetes to produce a variety of bioactive substances has been known for long time and being utilized in research and synthesize bioactive compound in numerous academic and industrial laboratories. These studies have tremendous impact on application against many human infections.14–16 Antibiotics resistance against infectious diseases has increased in recent years.17 Antibiotic resistance of pathogens (e.g. Staphylococcus aureus) causes a dangerous threat to the treatment of human infections. Scientists and medical professionals are worried about the potential threat. They are pursuing new experiments to discover unique and novel antibiotics against drug resistant pathogens.18,19 We have attempted to identify antibiotic producing bacteria from soil samples in fish and vegetable dump areas from Dhaka, Bangladesh. We have studied the biochemical and morphological properties of these bacteria. In this study, we report identification of three bacterial isolates producing antibacterial compound(s). Characterization of antibacterial factor(s) may lead to the discovery of novel antibiotic(s) with clinical application.
Chemicals and reagents
Trypticase soy broth (Hi-Media, India), Trypticase soy agar (Hi-Media, India), crystal violet, Gram’s iodine, safranin, ethyl alcohol, commercial antibiotic disc (Hi-Media, India).
Sample collection
Soil samples were collected from fish, vegetables and fruits dump area from Dhaka City.
Bacterial culture
About five grams of soil sample were suspended in TSB medium. The microbial suspension was further diluted (10-4) with the same medium. About100 µl of microbial suspension was spread on TSA plate and incubated for 24 hours. The bacterial colonies were isolated and grown in TSB. Bacteria were characterized.
Gram staining
A smear of bacterial cells was prepared on a clean glass slide and the smear was then allowed to air-dry followed by a mild heat fixation. Crystal violet solution was added onto bacterial smear and incubated for one minute. The smear was washed with water. Mordant Gram’s iodine solution was then added onto bacterial smear and incubated for one minute. The smear was decolorized by washing with 95% ethyl alcohol and rinsed with water. Finally safranin was used as counterstain for one minute and washed with water. Cells were then air dried and studied under microscope.
Antibacterial activity of soil isolates of bacteria
Bacterial strains isolated from soil samples were studied to determine their ability to kill other bacteria. The test was performed against E. coli, and S. aureus. E. coli or S. aureus were spread on separate TSA plates. The soil bacterial isolates were then inoculated as small spot with appropriate marking. The plates were then incubated at 37oC for 24 hours. Bacteria which can kill or produce chemicals that inhibit bacterial (E. coli, S. aureus, S. paratyphi, Acenobacter, V. cholerae and S. dysentery) growth will exhibit clear transparent zones around the inoculated spots of inoculation. After 24 hours of incubation, zone diameter was measured and recorded.
Identification of bacteria in soil samples from Dhaka city area
Soil around fish and vegetable waste dump area is an excellent environment for microbial growth due to abundant supply of organic material. In addition, warm temperature in Bangladesh favors bacterial growth. Hence we have selected and studied bacterial population in different food waste dumping area of Dhaka city. Soil samples were collected from fish and vegetable waste dumping area of Dhaka city. Soil samples were suspended in PBS and diluted to 10-4. A portion of diluted liquid was spread on to TSA plates. Bacteria were allowed to grow in a 37oC as shown in a sketch of procedure (Figure 1). After 24 hours of incubation, growth of bacteria was studied. Bacterial colonies were identified based on their color and morphology (data not shown). Single colonies were picked up and cultured in TSB for further analysis. Bacteria observed in this study were of different types as observed by morphology, color, and edge of colonies. The result obtained in this study is consistent with the previous reports.20,21
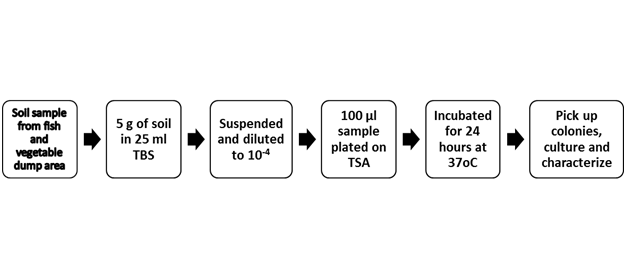
Figure 1 A flow chart showing the summary of isolation of bacteria from soil samples. Bacterial colonies were picked up as shown in the figure above. Individual colonies were cultured and used for characterization.
In recent years new bacterial pathogens are being found to be emerging with resistance to almost all antibiotics. It has become essential to develop novel antibiotics to overcome forth coming challenge of treating infectious diseases.22,23 We were interested if any of the soil bacterial isolates has the ability to produce chemicals that inhibit other bacterial growth. We inoculated a small dot of soil bacterial isolates on top of TSA plate pre-inoculated with E. coli, or S. aureus. Upon incubation for 24 hours, it was found that some of our bacterial isolates inhibit growth of both gram positive and gram negative bacteria (Figure 2A,2B). Interestingly K1, and K2 exhibited strong inhibitory effect on the growth of some pathogenic organisms (S. paratyphi and Acenobacter) (Table 1). Similarly K19 showed strong growth inhibitory activity against pathogenic bacteria, V. cholerae and S. dysentery (Table 1).
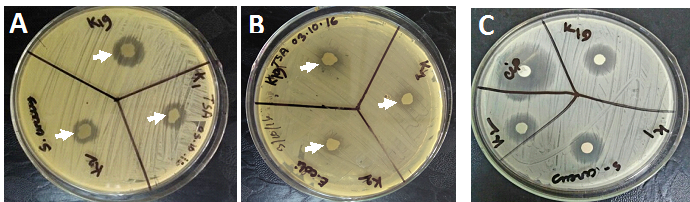
Figure 2 Bactericidal activity of soil bacterial isolates. Each of the bacterial isolates (K1, K2, and K19) were studied for their ability to produce bactericidal chemicals. S. aureus (A) or E. coli(B)cultures were inoculated on TSA plate by spreading with cotton swabs. A small amount of bacterial isolate was then spot inoculated on indicated position. TSA plates were then incubated for 24 hours and studied for the ability of soil bacterial isolates to inhibit growth of S. aureus(A) and E. coli (B). Clear zone (shown by arrow) was measured. C. Inhibitory effect of culture supernatant. Bacterial isolates (K1, K2, and K19) from soil samples were cultured in TSB for 24 hours. TSB culture was centrifuged at 12 krpm for 30 min. The supernatant was collected and filtered through 0.22 µm filter. About 50 µl of filtrate was soaked onto 3MM paper discs. Discs were applied onto pre-inoculated S. aureus TSA plate. The plate was incubated at 37oC overnight and studied for antibacterial activity. Arrows show clear zone where S. aureus did not grow. Ciprofloxacin (5 µg per disc) used as a positive control.
Bacterium |
Zone of Inhibition (Mm) |
||||||
S. aureus |
B. subtilis |
E. coli |
S. Paratyphi |
Acenobacter |
V. cholerae |
S. dysentery |
|
K1 |
20 |
18 |
>20 |
>30 |
>30 |
0 |
0 |
K2 |
10 |
6 |
>20 |
>30 |
>30 |
0 |
0 |
K19 |
30 |
ND |
20 |
>10 |
0 |
20 |
20 |
Table 1 Antibacterial activity of soil bacterial isolates. Each of the bacterial isolates (K1, K2, and K19) was studied for their ability to produce bactericidal chemicals. Individual test organism (listed in the Table) was inoculated on TSA plate by spreading with cotton swabs. A small amount of bacterial isolate was then spot inoculated on indicated position. TSA plates were then incubated for 24 hours and studied for the ability of soil bacterial isolates to inhibit growth of test organisms. Clear zone was measured and recorded. Experiment was repeated three times showing similar results
We studied whether antibacterial compound(s) diffuses out of the cell of soil bacterial isolates. Bacterial cultured medium was studied by disc diffusion method. Culture supernatant from K1, K2, and K19 inhibited growth of S. aureus (Figure 2C). Ciprofloxacin (5 µg per disc) was used as a positive control. Number of antibacterial compounds produced by these bacteria and the quantity of these compounds in the bacterial culture supernatant remain unknown. Therefore, a direct comparison between effect of ciprofloxacin and antibacterial compounds was impossible. We have heated the culture supernatant for 5 minutes and found no loss of its antibacterial activity (data not shown). Further studies are in progress to isolate and characterize antibacterial compounds produced by K1, K2, and K19. Once purified, the effect of antibacterial compounds produced by K1, K2, and K19 will be compared with commercially available antibiotics.
Bacterial isolates from soil samples of Dhaka city area are short rod-shaped cell
Soil bacterial isolates were studied by gram staining. It was found that K1 and K2 were gram negative, while K19 was gram positive (Figure 3A). Cellular morphology of all three (K1, K2, and K19) soil bacterial isolates was studied by scan electron microscopy. As shown in Figure 3B all three bacterial isolates were of small rod-shaped (coccobacilli).

Figure 3A Gram staining of bacterial isolates. Each bacterial sample (K1, K2, or K19) were spreaded on a glass slide and gram staining was performed as described in “Methods and Material”. Bacterial sample was studied under microscope and pictures were captured.
B. Bacterial isolates (K1, K2, or K19) were spreaded on a small cover slip. Morphology of bacteria was studied under a scan electron microscope and images were captured. Arrow shows position of individual cell.
Tolerance of bacterial isolates to environmental factors
All three bacterial isolates were studied for their tolerance to temperature, salt and pH. It was found that K1, K2, and K19 could grow between 20oC and 45oC (Table 2). All three bacteria could grow at pH range of 6-11 as well as NaCl concentration of 1-12% (data not shown). It remained to study further the enzyme producing ability of these bacterial isolates. In addition molecular analysis such as rRNA sequence analysis needs to be completed to determine the identity of each of these bacterial isolates.25
Bacteria |
4°C |
20°C |
37°C |
45°C |
60°C |
70°C |
K1 |
- |
+ |
+ |
+ |
- |
- |
K2 |
- |
+ |
+ |
+ |
- |
- |
K19 |
- |
+ |
+ |
+ |
- |
- |
Table 2 Effect of temperature on growth of bacterial isolates. One loopful of fresh bacterial cultures were inoculated into respective 5 ml TSB in test tube and incubated at 4oC, 20oC, 37oC, 45oC, 60oC and 70oC for 24 h. Turbidity of the culture media was determined after 24 h and indicated by + (Growth) and – (no growth) sign
This study describes isolation of three soil bacteria from Dhaka city area. Bacterial isolates were found to produce antibacterial components against both gram positive and negative cells. Bacterial cell morphology was studied by scan electron microscopy and found to be short rod shaped. Antibacterial compound(s) was found to diffuse out of the cell. Further studies are in progress to determine chemical nature of the antimicrobial compound(s).
The authors are thankful to the Department of Pharmacy, ASA University, Bangladesh for providing the necessary facilities to carry out the research work of this study. The work also was supported by the NIH-RISE (5R25GM105553-04).
There is no conflict of interest.
None.

©2017 Begum, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.