Journal of
eISSN: 2373-437X


Research Article Volume 3 Issue 1
Department of Biotechnology, Dr. Y. S. Parmar University of Horticulture and Forestry, India
Correspondence: Ambika Verma, Ph.D Scholar, Department of Biotechnology, Dr. Y. S. Parmar University of Horticulture and Forestry, Nauni, Solan-173230 (H.P.), India
Received: October 25, 2015 | Published: January 11, 2016
Citation: Verma A, Dhiman K, Shirkot P (2016) Hyper-Production of Laccase By Pseudomonas putida LUA15.1 through Mutagenesis. J Microbiol Exp 3(1): 00080. DOI: 10.15406/jmen.2016.03.00080
Pseudomonas putida LUA15.1, isolated from rice rhizospheric soil samples of paddy fields of Una district, Himachal Pradesh (India), produced extracellular laccase (EC 1.10.3.2) at 28°C and pH 7.0. In this study the wild strain of Pseudomonas putida LUA15.1was improved for enhanced laccase production by using physical mutagen (ultraviolet rays) and chemical mutagens (ethidium bromide (0.1-1.0mg/ml) and ethyl methanesulfonate (10-100µl/ml)). Mutant (E4) with hyper laccase production was obtained after treating wild strain with ethyl methanesulfonate on the basis of zone formation and laccase activity. Mutant E4 (34.12 U/l) resulted in a 26.37% increase in laccase activity over wild strain (25.12 U/l). Pseudomonas putida LUA15.1 mutant E4 exhibited maximum extracellular laccase activity at pH: 8.0, after 48hrs of incubation period at 30°C. The laccase enzyme was purified from Pseudomonas putida LUA15.1 mutant E4 by using ammonium sulfate precipitation, gel filtration chromatography and ion exchange chromatography. The procedure yielded 1.02 mg protein with 60.0 fold purification with a percent yield of 8.09. The effectiveness of hyper laccase producing mutant E4 indicates its possible applicability in various biotechnological and industrial processes.
Keywords: Pseudomonas putida 1, laccase, mutation, mutant
Laccases (benzenediol: oxygen oxidoreductases, EC 1.10.3.2) are multicopper blue oxidases that are widely distributed in higher plants, some insects, fungi and bacteria.1 Microbial enzymes continue to draw greater attention as alternative to chemical processes which would enable the industries to meet the increasingly stringent environment requirements to reduce the pollution load. The highly-stable bacterial laccases can function within a wider pH range and at high temperatures and these are less dependent on metal ions and less susceptible to inhibitory agents.2 Thus, bacterial laccases have significantly more potential industrial applications, especially in decolorization of industrial textile dye effluents which constitute a major threat to soil and ground water reservoirs worldwide. Furthermore, bacterial systems are easier to handle than fungal ones.3 Laccase activity has also been demonstated in a number of bacteria including Bacillus subtilis, Bacillus vallismortis, Bacillus pumilus, Bordetella compestris, Caulobacter crescentus, Escherichia coli, Mycobacterium tuberculosum, Pseudomonas putida, Pseudomonas syringae, Pseudomonas aeruginosa, Yersinia pestis and Geobacillus thermocatenulatus.4‒11
Laccases show significant biotechnological potential in various industrial applications, including kraft pulp and dye bleaching, polymer synthesis, beverage stabilization, anticancer drug development and the reduction of environmental pollution generated by waste effluents.12‒14 However, industrial applications of laccases are usually hindered by long fermentation period, low laccase yield, as well as poor enzyme stability.15 In addition to enzyme source viz., microorganism and substrate, other factors should be considered to develop efficient enzymatic bioprocesses and to satisfy commercial needs. The current study focuses on mutation breeding, in which a mutant strain with high laccase productivity and enhanced enzyme properties was obtained. Traditional mutation and selection techniques have been exploited for the improvement of bacterial cultures and to enhance laccase production level in microbes. Traditionally, strain improvement involves mutagenesis of a suitable strain followed by random screening of mutants. This approach has been successful and has benefited in recent years from the application of important advances in science and technology.16 An important such technique, random mutagenesis, is used to induce mutation in organisms using ultraviolet (UV), ethyl methyl sulfonate and ethidium bromide for better distinctiveness.17 The UV-induced mutagenesis is a frequently used method for breeding. The pyrimidine bases have strong UV absorption. When UV ray is absorbed, the neighboring double thymines in the DNA chain form a thymine dimer that mainly causes the mutations. When DNA reproduction begins, the site of the single thymine dimer could run over and then the gap is left, thereby inserting the false bases, causing the thymine-guanine (AT) → cytosine-adenine (GC) base pairs mutation.18 UV-induced mutagenesis produces point mutations, small deficiencies, chromosome breaks and chromosome rearrangements at different relative frequencies in many organisms.19 Quantitative enhancement is the foremost approach which involves strain improvement mostly by physical and chemical mutations. In recent years, many studies aimed to improve the strains using physical and chemical mutagenesis. Chemicals such as ethyl methanesulfonate, N-methyl-N′-nitro-N-nitrosoguanidine20 and physical such as UV have been commonly used.20,21
Therefore, the current study focuses on mutation breeding, aimed to enhance laccase production by improving the strain of Pseudomonas putida LUA15.1, in which the mutant strain with high laccase productivity and enhanced enzyme properties was obtained followed by partial purification of laccase.
Microbial strain
The microbial culture used in study was Pseudomonas putida LUA15.1, which was isolated from rice rhizospheric soil samples of paddy fields of Una district, Himachal Pradesh (India) and maintained on slants of nutrient agar medium at 4°C.
Culture conditions and enzyme extraction
Pseudomonas putida LUA15.1 was inoculated into the Tryptone Yeast (TY) broth and mixed thoroughly by keeping the flasks on a rotary shaker at 150rpm for 24-48 hours at 28°C. The culture supernatant was obtained by centrifugation of overnight culture of Pseudomonas putida LUA15.1 at 10,000rpm, for 10 minutes at 4°C and used for the enzyme assay.
Mutation of wild strain of Pseudomonas putida LUA15.1
Ultraviolet (UV) mutagenesis: For UV mutagenesis 24hr old culture was exposed to short Ultraviolet light (280 nm) from a distance of 15 cm for various time intervals (20, 40, 60, 80, 100 and 120 minutes). Sample was withdrawn before exposure to UV irradiation for determining the initial population (cfu/ml) on TY medium and similarly survival count after different time intervals was also determined. Survival curve was prepared and time of exposure giving 3 log kills was used for the selection of mutants.
Ethidium Bromide (Et Br) mutagenesis: The sample from 24hr old culture was used to determine the initial population (cfu/ml). Et Br (0.1-2.0mg/ml) was mixed well with culture in different test tubes at room temperature. The sample from each test tube was removed after an interval of 60 minutes for determining survival count (cfu/ml). The concentration resulting in 1 percent survival was used for carrying out mutation at 20, 40, 60, 80, 100 and 120 minutes interval in order to optimize the time period that would result in 3 log kill and survival curve was prepared.
Ethyl methanesulfonate (EMS) mutagenesis: One ml of 24hr old culture was centrifuged at 1000rpm for 2 minutes. The pellet was dissolved in Na-PO4 buffer (1ml, pH 6.5). 100µl of sample content was withdrawn to determine initial population (cfu/ml). EMS solution (10-100µl/ml) was added to the sample in different Eppendorf tubes and a concentration resulting in 1% survival was selected after an interval of 20min. The selected concentration was used for carrying out mutation at 20, 40, 80, 100 and 120 minutes interval in order to optimize the time period that would result in 3 log kill and survival curve was prepared.
Screening of mutants
Plate assay is a rapid determination for the screening of mutants with difference in laccase activity using tannic acid agar plate test. Tannic acid agar plate test involved the addition of 0.5% tannic acid into TY agar medium. Bacterial isolates were inoculated on this medium and plates were incubated for 3-4 days at an optimum incubation temperature of 28°C. Bacterial colonies which were able to produce maximum black zone with respect to wild strain were selected to be positive mutants for laccase activity and were further selected based on their capacity of enzyme production in liquid medium.
Statistical analysis
Complete randomized design (CRD) was used for statistical analysis of enzyme activity of wild as well as mutant strains.
Laccase production and extraction
Pseudomonas putida LUA15.1 and its mutant was inoculated into the Tryptone Yeast (TY) broth and mixed thoroughly by keeping the flasks on a rotary shaker at 150rpm for 24-48 hours at 28°C. The culture supernatant was obtained by centrifugation of overnight cultures at 10,000rpm, for 10 minutes at 4°C and used for the enzyme assay.
Laccase Activity Assay
Laccase activity was measured by monitoring the oxidation of ABTS.22 Catalase was added to the assay solution and incubated for 1 hour at 37°C to remove the possible effect of H2O2 produced by the bacteria. Laccase activity was determined spectrophotometrically at 420nm with ABTS as a substrate. The reaction mixture contained 200µl aliquots of crude extracellular enzyme preparation and 0.2mM ABTS in 0.1M sodium acetate buffer (pH 4.5) making final volume to 1.0ml. The reaction was held at 32°C for 10 mins followed by addition of 0.5ml of 80% trichloroacetic acid to stop the reaction. One unit of enzyme was defined as the amount of enzyme required to oxidize 1.0µmol of ABTS per min. The molar extinction coefficient of ABTS was found to be 36,000 M-1 cm-1. On the basis of maximum laccase activity, hyper laccase producing mutant of Pseudomonas putida LUA15.1, was selected for further studies.
Optimization of culture conditions for maximum laccase production
The mutant of Pseudomonas putida LUA15.1 selected on the basis of maximum laccase activity has further investigated to study the effect of different factors such as incubation temperature, pH and incubation time on laccase production as well as growth of the bacterial isolate. The pH range was optimized using TY medium adjusted from 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0 and 10.0 separately and temperature range for incubation investigated varied from 20-50°C where as effect of different incubation times for the growth of these thermophilic bacteria was studied for 24, 48, 72, 96, 120 and 144 hrs. In all cases, optical density was monitored by using a double beam UV/VIS scanning spectrophotometer.
Purification of laccase enzyme
All steps of purification were performed at a temperature of 4°C using 100 mM sodium phosphate buffer, pH 6.5. Techniques used for the purification of laccase enzyme were ammonium sulfate precipitation, dialysis, gel filtration chromatography and ion exchange chromatography. The enzyme preparations at various stages of laccase enzyme purification were analyzed for protein concentration and enzyme activity.
Ammonium sulfate precipitation: The cell free extract was subjected to various saturation/concentration of ammonium sulfate (0-90%). The ammonium sulfate precipitation table,23 was followed to calculate the required amount of ammonium sulfate to be added in cell free extracts. Ammonium sulfate was added with continuous stirring and stored at 4°C for overnight. The precipitated proteins were recovered by centrifugation at 20,000g for 20mins and the pellet was dissolved in minimum quantity of 100mM sodium phosphate buffer (pH 6.5). The dialysis was carried out for 24hrs against three successive changes of dialysis buffer 100mM sodium phosphate buffer (pH 6.5) in dialysis bag. The fraction or percentage cut exhibiting maximum activity of laccase enzyme was termed as ASF (Ammonium Sulphate Fraction) and was taken for further purification of laccase enzyme.
Gel filtration column chromatography: After the suspension obtained by ammonium sulphate precipitation and dialysis, enzyme solution was applied to Sephadex G-100 column (120cm×1cm), which has been pre-equilibrated with 100mM of phosphate buffer (pH:6.5) and later the enzyme has been obtained with the very same buffer. All of fractions have been analyzed for protein concentration and enzyme activity and the active fractions have been combined, concentrated and allowed to stand at 4°C.
DEAE Anion Exchange Chromatography: DEAE column (30x2.5 cm) was loaded with pooled sample from the previous step and equilibrated with 100mM of phosphate buffer (pH: 6.5) to wash out unbound proteins. The bound proteins were eluted with linear salt gradient using four bed volumes of 0.1M NaCl, 0.2M NaCl, 0.3M NaCl and 0.4M NaCl in phosphate buffer (pH 6.5). Fractions of 2ml each were collected and monitored for protein content at 280nm and for enzyme assay. The active fractions were pooled together and concentrated by dialysis against solid sucrose and used for further studies.
In this study, the yield of laccase enzyme from Pseudomonas putida LUA15.1 was increased by mutagenesis. Mutagens lead to mutation by inducing lesion or modification in the base sequence of DNA that remained un-repaired. Thus, an attempt was made to develop Pseudomonas putida LUA15.1 by mutagenesis and selection of efficient strain for production of laccase enzyme. The parent strain was treated with UV irradiations, Et Br and EMS.
Screening and selection of mutants by mutagenesis
UV mutagenesis: The survival count at each interval (20, 40, 60, 80, 100 and 120 minutes) was determined. Figure 1a, showed that 99.75% of the cells were killed within 120 minutes exposure of UV rays. Six putative UV mutants of Pseudomonas putida LUA15.1 were plated onto Tryptone Yeast (TY) medium containing 0.5% tannic acid to determine the size of clear zone. Of the 6 putative mutants which showed variable zones as compared to wild strain (0.5cm) when examined for production of laccase in liquid medium, Pseudomonas putida LUA15.1 mutant UV2 was selected with zone of 0.8cm and were found to produce 26.96 U/l of laccase activity in liquid medium as compared to wild type which yielded 25.12 U/l. Thus showing increase in laccase production by 6.82%.
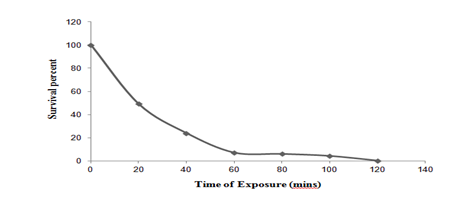
Figure 1a Survival curve of Pseudomonas putida LUA15.1 after UV irradiation at different time interval.
Et Br mutagenesis: Culture of Pseudomonas putida LUA15.1 was treated with different concentration of Et Br (0.1-1.0 mg/ml) for 60 minutes. 1mg/ml of Et Br resulted in 1.25 % survival and this concentration was further evaluated at different time interval (0 to 120 min). After 120 minutes of Et Br exposure (1 mg/ml) 99.90 % kill was recorded (Figure 1b). Survivors were observed for any change in the size of clear zone. Six putative mutants were obtained by Et Br mutagenesis, which were studied for laccase production in liquid medium. Of all the putative mutants tested, mutant EB2, EB3 and EB4 were selected with laccase activity index of 1.3, 1.5 and 2.2cm respectively, and were accounted for 7.13%, 12.32% and 21.86% increase in laccase activity (27.05 U/l, 28.65 U/l and 32.15 U/l respectively) in liquid medium as compared to wild strain (25.12 U/l).
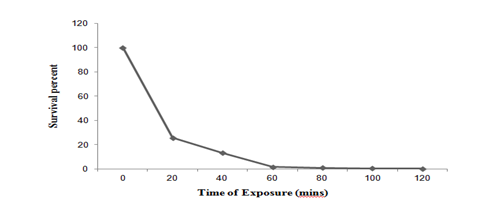
Figure 1b Survival curve of Pseudomonas putida LUA15.1 after exposure with Ethidium Bromide (1mg/ml) at different time interval.
EMS mutagenesis: In the present study, effect of different concentration of EMS (10-100µl/ml) on Pseudomonas putida LUA15.1 for 20 minutes was evaluated. Perusal of Figure 1c reveals that 100µl/ml) resulted in 0.94% survival because increase in EMS concentration (10-100µl/ml) declined the percentage of survivors, while the mutant frequency increased. Therefore, this concentration was further evaluated at different time interval (0 to 120 min). After 120 minutes of EMS exposure (100µl/ml) 99.95 % kill was recorded. The treatment of Pseudomonas putida LUA15.1 with 100µl/ml of EMS for 120 minutes resulted in 6 putative mutants on the basis of size of clear zone. These putative mutants were allowed to grow in liquid medium for enzyme production. Mutagenesis using EMS mutagen posed a significant increase in enzyme activity. Of all the putative mutants tested, mutant E2, E3, E4, and E5 was selected with the clear zone of 1.5, 1.7, 2.3 and 2.1cm respectively, and were accounted for 8.52%, 12.25%, 26.37% and 21.45% increase in laccase activity (27.46 U/l, 28.63 U/l, 34.12 U/l and 31.98 U/l) respectively in liquid medium as compared to wild strain (25.12 U/l).
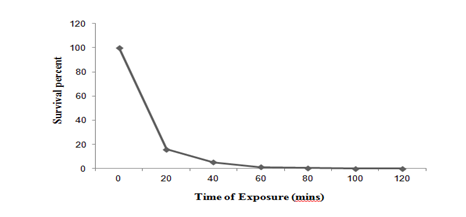
Figure 1c Survival curve of Pseudomonas putida LUA15.1 after exposure with Ethyl Methanesulfonate (100µl/ml) at different time interval.
UV, Et Br, and EMS resulted into 18 putative mutants, out of which 8 mutants were further selected on the basis of maximum laccase production in liquid medium, i.e. UV2, EB2, EB3, EB4, E2, E3, E4 and E5 (Figure 2). Laccase production by wild and mutant strains of Pseudomonas putida LUA15.1, revealed that maximum laccase activity was observed for mutant E4 (34.12 U/l) which was 26.37% higher than wild strain whereas other mutant strains [UV2 (26.96 U/l), EB2 (27.05 U/l), EB3 (28.65 U/l), EB4 (32.15 U/l), E2 (27.46 U/l), E3 (28.63 U/l) and E5 (31.98 U/l) resulted in 6.82%, 7.13%, 12.32%, 21.86% 8.52%, 12.25% and 21.45% increase in laccase activity, respectively (Table 1, Figure 3). The UV mutant, UV2 of Pseudomonas putida LUA15.1 illustrated 6.82% increase in enzyme activity. Increased laccase activity for mutant UV2 could be because of the reason that the gene responsible for production of laccase might have increased in DNA due to mutation (this is due to increase in gene copy number or expression of genes or both) which resulted in increase of enzyme activity. Similarly, laccase production has also been improved through UV radiations based mutagenesis, and UV mutant i.e. GZ11K2 was reported as a preferable laccase producer.21 Likewise, the chemical mutagens are stronger mutagenic agents and cause permanent changes in DNA sequence. They bring about transitions from G:C"A:T and have a preferential effect on DNA replication. Chemical mutagen used in the present study were Et Br and EMS. Et Br mutants EB2, EB3 and EB4 resulted in 7.13%, 12.32%, and 21.86% increase in laccase activity, respectively. Similarly, Ethidium Bromide has also been reported to increase laccase activity in fungi and bacteria.24,25 EMS resulted in mutants exhibited 8.52%, 12.25%, 26.37% and 21.45% improvement in enzyme activity respectively. Similarly, the effect of EMS on laccase production by Streptomyces lydicus has also been reported.26 On the basis of maximum laccase activity, Pseudomonas putida LUA15.1 mutant E4 (34.12 U/l), which was 26.37% higher laccase producer than wild strain, was selected for further studies.
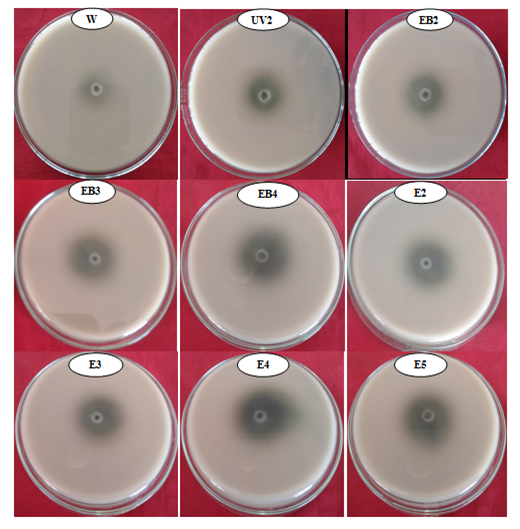
Figure 2 Selected eight mutants of Pseudomonas putida LUA15.1 on the basis of maximum laccase production i.e. UV2, EB2, EB3, EB4, E2, E3, E4 and E5.
|
Strains of Pseudomonas putida LUA15.1 |
||||||||||
|
|
WILD |
UV2 |
EB2 |
EB3 |
EB4 |
E2 |
E3 |
E4 |
E5 |
CD0.05 |
|
Zone (cm) |
0.5 |
0.8 |
1.3 |
1.5 |
2.2 |
1.5 |
1.7 |
2.3 |
2.1 |
2.8 |
|
Activity (U/l) |
25.12 |
26.96 (6.82↑) |
27.05 (7.13↑) |
28.65 (12.32↑) |
32.15 (21.86↑) |
27.46 (8.52↑) |
28.63 (12.25↑) |
34.12 (26.37↑) |
31.98 (21.45↑) |
|
Table 1 Laccase production by Pseudomonas putida LUA15.1 and its selected mutants
Statistical analysis: Complete randomized design (CRD) was used for statistical analysis of laccase enzyme activity of wild as well as its mutant strains. Maximum laccase activity was observed for mutant E4 (34.12 U/l) which was statistically at par with the laccase enzyme activities of two mutants viz., E5 (31.98 U/l) and EB4 (32.15 U/l) whereas minimum laccase enzyme activity was observed for wild strain of Pseudomonas putida LUA15.1 which was statistically at par with the laccase enzyme activities of three mutants viz., UV2 (26.96 U/l), EB2 (27.05 U/l) and E2 (27.46 U/l).
Optimization of culture conditions for maximum laccase production
Pseudomonas putida LUA15.1 mutant E4 was selected for the optimization of best growth conditions with the objective to produce maximum laccase enzymes. Each organism or strain has its own special conditions for the maximum enzyme production. The general rules for the optimization of microbial enzyme production have been affected by various physical factors which include pH, temperature and time. These factors have been found to be important to promote, stimulate, enhance and optimize the production of enzymes. The isolate exhibited maximum growth OD of 1.45 at a wavelength of 540nm after 96hrs, whereas maximum extracellular activity of 33.80 U/l was observed after 48hrs of the incubation period. In other reports, γ-proteobacterium JB produced maximum intracellular as well as an extracellular laccase activity after 48hrs of incubation, whereas maximum laccase production of 94.10 U/ml was recorded in Pseudomonas putida MTCC 7525 after 108hrs of incubation.27 The selected bacteria showed a maximum growth OD of 1.632 and maximum extracellular activity of 33.70 U/l at an incubation temperature of 30°C. In other studies of laccase production it was noted that, maximum enzyme production was achieved at 30°C temperature in case of Pseudomonas putida MTCC 752527 and at 37°C in case of γ-proteobacterium JB.28 Maximum growth OD of 1.51 and maximum extracellular activity of 34.30 U/l was observed at pH: 8.0. Maximum enzyme production has been reported in other studies at an optimum pH of 8.0 such as in case of Pseudomonas putida MTCC 752527 and γ-Proteobacterium JB.28
Purification of laccase enzyme from Pseudomonas putida LUA15.1 mutant E4
Crude extracellular enzyme extract was subjected to ammonium sulphate (0-90%) saturation and it was found out that maximum laccase enzyme activity of 32.22 U/l was detected in 50-90 percent level of saturation. The enzyme preparation at this stage was treated as ammonium sulphate fraction (ASF). The ASF (proteins precipitated after the overnight dialysis of 50-90% ammonium sulphate cut) exhibits the highest rate of precipitation and purified the enzyme 8.0 times with a 82.73% yield and was applied on Sephadex G-100 column equilibrated with sodium phosphate buffer (pH 6.5), purified the enzyme 29.03 times with a 31.62% yield. And in the last step, the enzyme fractions obtained and concentrated from Sephadex G-100 column were applied to the DEAE sephadex ion exchange column. It was found that laccase fractions were eluted with 1.0 M NaCl gradient which further purified laccase with 60.0 times purification and 8.09% yield. Laccase activity was calculated using the ABTS assay. All purification steps of the laccase enzyme are shown in Table 2. In other studies, laccases have also been purified from certain bacteria like Azospirillum lipoferum, Marinomonas mediterranea, Bacillus subtilis and γ-Proteobacterium JB.4,18,29
|
Steps |
Total Enzyme Activity (U) |
Total soluble protein (mg) |
Specific activity (U/mg protein) |
Fold purification |
Percent yield |
|
Crude extract |
2.346 |
755 |
0.0031 |
1 |
100 |
|
Ammonium sulphate precipitation |
1.941 |
78.12 |
0.0248 |
8 |
82.73 |
|
Gel filtration chromatography |
0.742 |
8.24 |
0.09 |
29.03 |
31.62 |
|
Ion exchange chromatography |
0.19 |
1.02 |
0.186 |
60 |
8.09 |
Table 2 Partial purification of laccase from Pseudomonas putida LUA15.1 mutant E4
In this study the wild strain of Pseudomonas putida LUA15.1 was improved for enhanced laccase production by using physical and chemical mutagens. Mutant (E4) with hyper laccase production was obtained after treating wild strain with ethyl methanesulfonate on the basis of the maximum zone formation and laccase activity. Mutant E4 (34.12 U/l) resulted in a 26.37% increase in laccase activity over wild strain (25.12 U/l) at pH: 8.0, after 48hrs of incubation period at 30°C. The laccase enzyme was purified from mutant E4 by using ammonium sulfate precipitation, gel filteration chromatography and ion exchange chromatography. The procedure yielded 1.02mg protein with 60.0 fold purification with a percent yield of 8.09. Mutant E4 resulted in 26.37% increase in laccase activity over wild strain. The mutant strain E4 of Pseudomonas putida LUA15.1 was more effective in producing laccase as compared to wild strain, hence it can be very useful in various biotechnological applications.
None.
Authors declare that there is no conflicts of interest.

©2016 Verma, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.