Journal of
eISSN: 2373-437X


Research Article Volume 2 Issue 4
Department of Medicine, Georgia Regents University, USA
Correspondence: Elias K Manavathu, Ph. D., Section of Infectious Diseases, Department of Medicine, Georgia Regents University, 1120 15th Street, Room AE 3033, Augusta, GA., 30912, USA, Tel 706-723-4159, Fax 706-446-5739
Received: June 15, 2015 | Published: September 22, 2015
Citation: Manavathu EK, Vazquez JA. Effects of antimicrobial combinations on Pseudomonas aeruginosa-Aspergillus fumigatus mixed microbial biofilm. J Microbiol Exp. 2015;2(4):126-136. DOI: 10.15406/jmen.2015.02.00057
Objectives: Co-infection with P. aeruginosa and A. fumigatus is frequently seen in patients with cystic fibrosis. These microorganisms are known to produce biofilm both in vitro and in vivo. The biofilm-embedded microbial cells are frequently refractory to conventional antimicrobial therapy. The primary objective of this study was to evaluate the efficacy of several anti-pseudomonal antimicrobials such as cefepime, imipenem and ciprofloxacin individually and in pair-wise combinations with antifungal drugs on P. aeruginosa, A. fumigatus polymicrobial biofilms and compare the results with those obtained in monomicrobial biofilm.
Methods: Biofilms of P. aeruginosa and A. fumigatus isolates were grown in 24-well cell culture plates in Sabouraud’s dextrose broth at 35˚C. The activities of cefepime, imipenem and ciprofloxacin alone and in two-drug combination with voriconazole, posaconazole, amphotericin B and anidulafungin on monomicrobial and polymicrobial biofilms, as well as on planktonic cells were examined by CFU assay.
Results: Scanning electron microscopic studies showed that A. fumigatus produced firmly adherent mixed microbial biofilm with P. aeruginosa on Thermanox plastic coverslips with increased synthesis of extracellular matrix in the presence of the bacterial cells. The fungal hyphae in monomicrobial and polymicrobial biofilms were realigned during biofilm growth forming parallel-packed bundles with no apical or dichotomous branching. Typically, P. aeruginosa produced firmly adherent mixed microbial biofilm with increased synthesis of extracellular matrix in the presence of A. fumigatus hyphae, but tended to form loosely adherent monomicrobial biofilm. Overall, the susceptibility of P. aeruginosa to cefepime and imipenem alone and in pair-wise combination with antifungal drugs was significantly decreased in polymicrobial biofilms (»0 to 0.5 logs CFU reduction at 16µg/ml) when compared to monomicrobial biofilms (»1.5 to 4.5 logs CFU reduction at 16µg/ml) (p values ranged from 0.0076 to 0.0509). On the other hand, the efficacy of ciprofloxacin in monomicrobial and polymicrobial biofilms was similar (»2.5 to 3.5 logs CFU reduction at 16µg/ml). A. fumigatus monomicrobial and polymicrobial biofilms were similarly susceptible to antifungal drugs with and without the antibacterial in the combination. Time-kill experiments performed at 4 times the MICs of the drugs (0.5µg/ml to 4µg/ml) showed that the planktonic cells of P. aeruginosa (»4 to 4.5 logs CFU reduction) and A. fumigatus (»2.5 to 3 logs CFU reduction) in monocultures and mixed microbial cultures were similarly susceptible to antimicrobial drugs.
Conclusions: In our model, the P. aeruginosa cells associated with P. aeruginosa-A. fumigatus polymicrobial biofilms were recalcitrant to certain antibacterial drugs compared to monomicrobial biofilms, whereas the planktonic cell monocultures and mixed microbial cultures showed no significant difference in their antimicrobial drug susceptibility profiles.
Keywords: polymicrobial biofilm, drug combinations, differential activity, antimicrobial drugs, drug susceptibility
Application of metagenomics1‒5 and high-resolution culture independent molecular probing techniques6‒10 have revealed that the microbiota of the airways of cystic fibrosis (CF) patients is highly complex and consists of 100s of microorganisms belonging to a wide variety of taxonomic groups. The pulmonary function of CF patients is significantly affected by the interaction of these microorganisms in the respiratory tract and by the proinflammatory host immune response generated against the airway microbial community. Furthermore, application of conventional culture dependent methods have identified a core group of bacterial pathogens consisting of Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, Burkholderia cepacia complex, Alcaligenes xylosoxidans and Stenotrophomonas maltophilia,11‒13 colonizing/infecting the airways of CF patients in an age dependent manner. Haemophilus influenzae (≥16.3%) and S. aureus (≥50.9%) are highly prevalent in children and adolescents, whereas P. aeruginosa is frequently (prevalence ≥52.5%) isolated from adult CF patients.14,15
In addition to the bacterial species, the microbiota of CF airways often consists of fungi such as Aspergillus fumigatus,16,21 Candida species,22‒26 Scedosporium species,27‒30 Exophiala dermatitides31‒34 and Pneumocystis species,35‒38 mostly in combination with bacteria producing mixed microbial colonization/infections in the CF airways.39 The bacterial and fungal species more frequently isolated from the airway secretions of CF patients with mixed microbial colonization/infections are P. aeruginosa and A. fumigatus. These opportunistic pathogens frequently produce monomicrobial and polymicrobial biofilms both in vitro,40 and in vivo.41 Genesis, architecture and drug susceptibility of in vitro and in vivo monomicrobial biofilms of P. aeruginosa have been the subject of investigation over the past two decades.42‒44 It has been shown that the biofilm-bound P. aeruginosa cells are highly tolerant/resistant to antimicrobial drugs45‒50 compared to their planktonic counter parts. However, to date very little is known about the development, structure and the effectiveness of antimicrobial drug treatment against the P. aeruginosa-A. fumigatus (Pa/Af) polymicrobial biofilm complex produced by mixed cultures of these microorganisms. More recently, A. fumigatus in vitro and human bronchial epithelial cell culture monomicrobial biofilm models were described to study the susceptibility of biofilm-associated fungal cells to antimicrobial drugs.51‒53 We recently described an in vitro Pa/Af polymicrobial biofilm model for studying the antimicrobial drug susceptibility of this mixed microbial biofilm.40 Our preliminary study showed that the drug susceptibility of A. fumigatus to the triazoles such as voriconazole and posaconazole remained the same both in monomicrobial and polymicrobial biofilms, whereas the susceptibility of P. aeruginosa to cefepime was significantly reduced in polymicrobial biofilm compared to that of monomicrobial biofilm. However, the efficacy of tobramycin was unchanged in the polymicrobial biofilm vs monomicrobial biofilm. This difference in antibacterial activity against biofilms prompted us further study the antimicrobial susceptibilities of monomicrobial and polymicrobial biofilms of Pa/Af.
Thus, the primary objective of this study was to expand on our previous preliminary observations describing the differential susceptibilities of P. aeruginosa to cefepime in monomicrobial and polymicrobial biofilms by including additional antimicrobial drugs and isolates of P. aeruginosa and A. fumigatus. The key question was whether the formation of a mixed microbial biofilm provides added protection to either the bacterial or the fungal cell from the effects of antimicrobial drugs alone or in combinations when compared to the effects on monomicrobial biofilms produced by P. aeruginosa and A. fumigatus. The in vitro effectiveness of individual and pair-wise combinations of the drugs was examined by determining the reduction of CFU after a 24h exposure to the drug(s).
Microorganisms: P. aeruginosa isolates 56402 (PA56402: a mucoid clinical isolate obtained from the Microbiology Laboratory of Henry Ford Hospital in Detroit, MI, USA), PAO1 (obtained from Matthew Parsek, University of Washington, Seattle, WA, USA) and P. aeruginosa 27853 (PA27853: American Type Culture Collection, Manassas, VA, USA) were used in this study. The cultures preserved as freezer stocks in -80˚C were subcultured on Brain Heart Infusion (BHI) agar for the evaluation of purity and viability. Working cultures were routinely grown on BHI agar, stored at 4°C and sub-cultured once a week to maintain viable stock cultures. PA56402, PAO1 and PA27853 were susceptible to a variety of antibacterial drugs such as aminoglycosides, b-lactams and fluoroquinolones, including cefepime (MIC ≤1µg/ml), imipenem (MIC ≤2µg/ml) and ciprofloxacin (MIC ≤0.25 µg/ml). Sabouraud’s dextrose (SD) agar and SD broth were used for growing monomicrobial and mixed microbial cultures producing biofilms. One ml aliquots of the overnight cultures were centrifuged in a microcentrifuge at top speed for 2min and the pellets washed 3 times with sterile distilled water, resuspended in 1 ml fresh SD broth, standardized spectrophotometrically using a standard curve, and then used for the various experiments. The use of SD agar and SD broth was particularly convenient since these media were commonly used to grow A. fumigatus.
The A. fumigatus clinical isolates 53470 and 43135 (AF53470 and AF43135) obtained from the Microbiology Laboratory of Henry Ford Hospital in Detroit, Michigan, USA and ATCC 36607(AF36607) purchased from American Type Culture Collection were used in this study. The initial cultures were subcultured on SD agar to evaluate viability and purity of the culture, and subsequently stored as conidial suspension in 25% glycerol at -80°C. Working cultures were maintained on SD agar plates at 4°C. AF53470, AF43135 and AF36607 were all susceptible to polyenes, triazoles and echinocandins, including amphotericin B, voriconazole, posaconazole (MICs 1µg/ml, 0.25µg/ml, 0.125µg/ml, respectively) and anidulafungin (minimum effective concentration 0.031µg/ml). For preparation of conidia, cultures were grown on SD agar plates for 4 days at 35°C to produce conidia. The SD agar containing mycelial growth was cut into small (5 mm2) pieces using a sterile spatula, transferred to a 50-ml screw-capped conical culture tube containing 25 ml sterile distilled water and vortexed vigorously for 2 min to disperse the conidia from the conidiophores. The resulting fungal suspension was filtered through 8 layers of sterile cheese cloth to remove mycelial and agar debris. The clarified conidial suspension thus obtained was standardized by hemocytometer count and stored at 4˚C in the refrigerator.
Scanning electron microscopy: The monomicrobial and polymicrobial biofilms of A. fumigatus and P. aeruginosa were grown on sterile tissue culture Thermanox 13mm plastic coverslips (Nalgene Nunc International, Rochester, NY, USA) in SD broth at 35°C. For the development of polymicrobial biofilm, sterile plastic coverslips were placed in 12-well Costar tissue culture plate and 2ml A. fumigatus conidial suspension (1x106 conidia/ml) was placed in each well completely covering the submerged plastic cover slips. The tissue culture plate with the cover slips was incubated statically at 35°C for 18h for the conidia to germinate and form a monolayer of mycelial growth on the plastic coverslips. The spent growth medium from each well was removed and the mycelial growth was washed 3 times with sterile distilled water (2ml each) and inoculated with 2ml SD broth containing 1x106 P. aeruginosa cells/ml. The mixed culture was incubated for 24 h at 35°C for the development of polymicrobial biofilm. The plastic cover slips containing the mixed microbial growth were washed 3 times with sterile distilled water (2ml each) and transferred to a clean 12-well Costar tissue culture plate and the biofilm was fixed for 60 min in 2% glutaraldehyde in 0.1M sodium cacodylate (NaCac) buffer (pH 7.4), postfixed in 2% osmium tetroxide in NaCac buffer, dehydrated with a graded ethanol series (25-100%) and critical point dried in CO2. The dried specimens were mounted on aluminum stubs with carbon adhesive tabs and sputter coated with gold-palladium. Biofilm was observed and imaged in a FEI XL30 scanning electron microscope (FEI, Hillsboro, OR) at 10 kV. For the development of monomicrobial biofilms A. fumigatus and P. aeruginosa, monocultures of these organisms were grown on Thermanox coverslips for 24h at 35˚C, washed and processed for SEM as described above. The images were edited and processed using SPOT image processing computer software.54
Determination of the effects of antibiotics on biofilms: A. fumigatus conidia (1x106 conidia/ml) were grown in 1ml SD broth in Costar 24-well cell culture plates at 35°C for 18h. The surface growth was removed and the adherent mycelial growth on the bottom of the plastic well was washed 3 times with sterile distilled water (1ml each) and inoculated with 1x106 P. aeruginosa cells in 1ml SD broth, and incubated at 35°C for 24h for the development of polymicrobial biofilms. Monocultures of P. aeruginosa and A. fumigatus were grown under identical conditions for the formation of monomicrobial biofilms. The biofilms were washed with distilled water (3 times, 1ml each) and incubated with the required concentrations of antimicrobial drug(s) for 24 h at 35°C. The drug-treated biofilms were washed and the adherent cultures containing either fungal or bacterial or a mixed population of fungal and bacterial cells were harvested by scraping the bottom of the wells of the cell culture plate using sterile swabs into 1 ml aliquots of sterile distilled water. The cell suspension was vortexed vigorously to disperse the cells, diluted 10 to 108 fold and 0.01 ml aliquots of the cell suspensions were plated on ciprofloxacin (50µg/ml) or voriconazole (16µg/ml) containing SD agar plates and incubated at 35°C for 24h for selective growth. The number of CFU for each group was determined and plotted against the drug concentration to assess the effectiveness of antibiotic treatment against biofilms.
In vitro susceptibility: The in vitro susceptibility of P. aeruginosa isolates to various antibacterial drugs listed in Table 1 was initially examined using the fully automated Vitek 2 system (bioMerieux, Inc., Durham, NC, USA) and subsequently confirmed for cefepime, imipenem and ciprofloxacin in our laboratory using CLSI Protocol M100-S19.55 Susceptibility testing with cefepime, imipenem and ciprofloxacin was repeated at least once and the results were identical.
|
Microorganism |
Antimicrobial drug |
MIC (mg/L) |
|
P. aeruginosa 56402 |
Amikacin |
≤2 |
|
Cefepime |
≤1 |
|
|
Ciprofloxacin |
≤0.5 |
|
|
Gentamicin |
≤1 |
|
|
Imipenem |
2 |
|
|
Piperacillin/tazobactam |
8 |
|
|
Tobramycin |
≤1 |
|
|
A. fumigatus 53470 |
Voriconazole |
0.25 |
|
Posaconazole |
0.125 |
|
|
Caspofungin |
0.031* |
|
|
Micafungin |
0.031* |
|
|
Anidulafungin |
0.031* |
|
|
Amphotericin B |
1 |
Table 1 In vitro susceptibility of Pseudomonas aeruginosa 56402 and Aspergillus fumigatus 53470 to various antibacterial and antifungal drugs
*Values represent minimum effective concentration.
The in vitro susceptibility of AF53470, AF43135 and AF36607 to voriconazole, posaconazole, amphotericin B and anidulafungin was determined by CLSI Document M38-A2.56 Drug concentrations ranging from 0.031µg/ml to 16µg/ml were used. The MICs of voriconazole, posaconazole and amphotericin B were defined as the lowest concentration of the drug that provided 100% growth inhibition whereas the effectiveness of the echinocandin was defined as the lowest concentration of the drug that produced a distinct morphological change (minimum effective concentration) resulting in granular appearance of the colony due to stunted mycelial growth. Each susceptibility test was repeated at least once and the results were either identical or ± one two-fold dilution.
Determination of bactericidal and fungicidal effects on suspension cultures: The fungicidal and bactericidal activities of various antifungal and antibacterial drugs alone and in two-drug combinations on monomicrobial and polymicrobial cell suspensions were examined by determining the microbicidal activity of the drug(s). Overnight cultures of P. aeruginosa isolates PA56402, PAO1 and PA27853 were grown in 5ml SD broth at 35˚C. The cells were collected by centrifugation, washed twice with sterile distilled water and standardized to 2x106 cells/ml in SD broth by a spectrophotometric method using a standard curve.
Sporeling suspensions of AF53470, AF43135 and AF36607 containing 2x106 sporelings/ml were grown for 12h at 35˚C were prepared in SD broth. One ml aliquots of monomicrobial and polymicrobial cell suspensions containing 1 x 106 cells each/ml were prepared by either 2-fold dilution of monomicrobial cell suspension or by mixing equal volumes of P. aeruginosa and A. fumigatus cell suspensions in Costar 24-well cell culture plates. Suspensions were then exposed to cefepime or ciprofloxacin alone or in two-drug combination with voriconazole, posaconazole, anidulafungin and amphotericin B at 4 times the MICs of the drugs (0.5µg/ml to 4µg/ml) for 24h at 35˚C with agitation (120rpm) on a gyratory shaker. The drug-exposed cells were collected, washed by centrifugation and resuspended in 1ml sterile distilled water. The cell suspension was diluted from 10 to 108 fold by ten-fold serial dilution. Subsequently, 0.01ml aliquots were plated on ciprofloxacin (50µg/ml) SD agar plates or on voriconazole (16µg/ml) containing BHI agar plates for selective growth and the CFU for each treatment group was determined.
Antimicrobial drugs: Cefepime (Sagent Pharmaceuticals, Schaumberg, IL, USA), imipenem and ciprofloxacin (Teva Parenteral Medicines, Inc., Irvine, CA, USA) were prepared into 1mg/ml stock solutions using sterile distilled water and stored as 0.25ml aliquots at -20˚C. The frozen stocks were thawed at room temperature and used within 24h. Antifungal drugs were obtained as pure powder from the manufacturers as follows: voriconazole and posaconazole were obtained from Pfizer Pharmaceuticals (New York, NY, USA) and Schering-Plough Research Institute (Kenilworth, NJ, USA), respectively. Amphotericin B was purchased from Sigma Chemical Company (St. Louis, MO, USA). Anidulafungin was obtained from Pfizer Pharmaceuticals, New York, NY, USA. The triazoles and amphotericin B were dissolved in DMSO to obtain a stock solution of 10mg/ml and stored as 0.25ml aliquots at -20˚C. Anidulafungin was dissolved in sterile double distilled water to obtain a concentration of 10 mg/ml and stored as 0.25ml aliquots at -80˚C. The frozen stocks of the antifungal drugs were thawed at room temperature and used within 24h. Where it is applicable, comparable concentrations of DMSO were used as control to examine its effect on the growth of the organism.
Statistical analysis: The data were analysed by Student’s t-test, one-way and two-way analysis of variance with Bonferroni’s Multiple Comparison Test using GraphPad Prism Version 5.0 for Windows (GraphPad Software, Inc., La Jolla, CA, USA). A p value ≤0.05 was considered significant. Details of each statistical test used are given in the corresponding figure legend.
Characteristics of monomicrobial and polymicrobial biofilms: Figure 1 shows scanning electron microscopic images of 24h monomicrobial and polymicrobial biofilms of A. fumigatus and P. aeruginosa. As shown in panel A, A. fumigatus conidia readily adhered to the Thermanox plastic coverslip and produced a confluent mycelial growth firmly attached to the plastic coverslip. Typically, lateral and apical dichotomous branching of hyphal filaments characteristic of A. fumigatus growing in rich medium appeared to be minimal and the hyphae extend by apical growth without branching. Bundles of parallel packed hyphal filaments (black arrow) held together by a fluffy extracellular matrix produced by the adhered hyphae (white arrows) were readily seen. In addition to the growing hyphae, ungerminated conidia and sporelings were also seen attached to the substrate as a result of non-synchronous spore germination and growth.

Figure 1 Scanning electron microscopic images of monomicrobial and polymicrobial biofilms of A. fumigatus and P. aeruginosa.
The biofilms were grown on13 mm sterile Thermanox plastic cover slips in SD broth in 12-well Costar cell culture plates and processed for electron microscopy as previously described. Panel A: Germinating A. fumigatus conidia produced a firmly adhered mycelial growth on the plastic cover slips immersed in SD broth at the bottom of the cell culture plate. The mycelial growth mainly consists of unbranched hyphae often forming bundles of parallel packed hyphal filaments (black arrow). The parallel packed hyphal filaments were held together by a fluffy extracellular matrix produced by the adhered hyphae (white arrows). Panel B: Light microscopic image of a growing region of an A. fumigatus colony grown in a shake suspension culture showing numerous hyphal branches. Representatives of apical dichotomous and lateral hyphal branches are indicated by the black arrows. Panel C: P. aeruginosa formed a multilayer aggregate of cells seemingly held together by extracellular matrix and perhaps by surface appendages such as flagella and surface pili (fimbriae) (white arrow) not only among themselves but also to the substrate. The monomicrobial biofilm produced by P. aeruginosa was loosely adhered to the plastic cover slips and often dislodged by vigorous agitation. The extracellular matrix partially covering the cell surface was visible (black arrow) in 24-h P. aeruginosa biofilm. Panel D: Static coincubation of A. fumigatus sporelings pregrown for 12 h or longer with P. aeruginosa cells produced a mixed community of cells consisting of fungal hyphae and bacterial cells. The bacterial cells were embedded in a mesh like extracellular matrix material and firmly adhered to the plastic cover slips. The extracellular matrix (white arrows) production was significantly enhanced and the bacterial cells held together by the surface appendages are also firmly attached to the fungal hyphae using it as support (black arrows) to build the mixed microbial biofilm community. The adherent bacterial cells were hardly removed by vigorous agitation during extensive washing of the biofilm.
Since one of the characteristic features of the monomicrobial biofilm of A. fumigatus was the absence of hyphal branching we examined the branching pattern of AF43135 (one of the A. fumigatus isolates used in this study) in SD broth in planktonic shake and static cultures at 35˚C. As shown in panel B, in the planktonic shake and static cultures AF43135 produced branches (arrows) extensively usually either at the junctional point where the septum formation takes place (lateral branches) or at the apex of the hypha (dichotomous branching) producing secondary and tertiary branches forming a prolific network of mycelial growth.
Figure 1 panel C shows the general architecture of monomicrobial biofilm of P. aeruginosa formed on Thermanox plastic coverslip. P. aeruginosa formed a multilayer aggregate of cells seemingly held together perhaps by surface appendages such as flagella and surface pili (fimbriae) (white arrow) and extracellular matrix not only among themselves but also to the substrate. The bulk of the monomicrobial biofilm produced by P. aeruginosa was loosely adhered to the Thermanox coverslips and was easily dislodged by vigorous agitation during extensive washing of the biofilm leaving a thin layer of cells firmly attached to the coverslips. The extracellular matrix partially covering the cell surface was visible (black arrow) in 24h P. aeruginosa biofilm.
Although the general appearance and architecture of the P. aeruginosa-A. fumigatus polymicrobial biofilm is similar to P. aeruginosa monomicrobial biofilm, the structure of the former was more complex. Inoculation of pregrown A. fumigatus mycelia adhered to Thermanox coverslip with P. aeruginosa cells produced a mixed community of cells consisting of fungal hyphae and bacterial cells (Figure 1 Panel D). The bacterial cells were embedded in a mesh-like extracellular matrix and firmly adhered to the coverslip using the fungal hyphae as scaffolding for firm adherence producing a prolific mixed microbial biofilm. Unlike the P. aeruginosa monomicrobial biofilm, the adherent bacterial cells in mixed microbial biofilm were not removed by extensive washing of the biofilm. The extracellular matrix (white arrows) production was significantly enhanced and the bacterial cells held together by the extracellular matrix as well as surface appendages were also firmly attached to the fungal hyphae using it as support (black arrows) to build the mixed microbial biofilm. In a 24h polymicrobial biofilm the matrix begins to encase the bacterial cells producing thicker biofilm. To evaluate the antibiotic susceptibilities of P. aeruginosa monomicrobial and Pa/Af polymicrobial biofilms we routinely used biofilms similar to the one shown in Figure 1.
Effects of antimicrobial drugs on biofilms: In vitro activities of cefepime, imipenem and ciprofloxacin individually and in two-drug combinations with four antifungal drugs, namely, voriconazole, posaconazole, amphotericin B and anidulafungin against monomicrobial and polymicrobial biofilms of Pa/Af were examined. Figure 2 shows the effects of voriconazole (panel A), posaconazole (panel B), amphotericin B (panel C) and anidulafungin (panel D) individually at 16µg/ml on A. fumigatus monomicrobial and Af/Pa polymicrobial biofilms. Among various antifungal drugs used posaconazole showed the best activity (2.59 to 2.64 logs CFU reduction) against A. fumigatus biofilms whereas voriconazole (1.19 to 1.37 logs CFU reduction), amphotericin B (0.92 to 0.98 logs CFU reduction) and anidulafungin (0.64 to 0.83 logs CFU reduction) produced moderate to minimum effects compared to the drug-free control (p values ranged from 0.0033 to 0.0303). Regardless of the antifungal drug used, both the monomicrobial and the polymicrobial biofilm-associated A. fumigatus cells were almost equally susceptible to the inhibitory effects of the antifungal drugs alone (p values ranged from 0.1382 to 0.8265) or in combination with the antibacterial drugs cefepime, imipenem and ciprofloxacin (data not shown). However, a comparison of the fungicidal activities of various antifungal drugs against suspension cultures (see section on planktonic assay) and biofilms (Figure 2) showed that the latter was substantially less susceptible to antifungal drugs compared to planktonic cells.
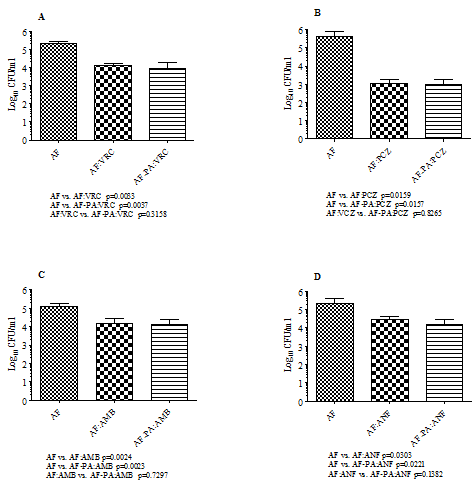
Figure 2 Antifungal activities of voriconazole (A), posaconazole (B), amphotericin B (C) and anidulafungin (D) on AF53470 monomicrobial and AF53470-PA56402 polymicrobial biofilms. The experiments were performed several times and the results shown here represent data from two independent experiments with three replications. Each histogram represents the mean of two independent experiments and the vertical bar on each histogram denotes the standard deviation. The data were analysed by pair-wise comparison of each group by Student’s t-test using Graphpad Prism 5.0 and a p value ≤0.05 was considered to be significant. The data shown in Figure 2 illustrate the activities of various antifungal drugs at a concentration of 16µg/ml. AF, A. fumigatus monomicrobial biofilm; AF-PA, A. fumigatus-P. aeruginosa polymicrobial biofilm; VRC, voriconazole; PCZ, posaconazole; AMB, amphotericin B; ANF, anidulafungin. In addition, the experiment was repeated twice with laboratory isolates AF36607 and PA27853 and similar results were obtained.
Figure 3 shows the effects of cefepime alone and in pair-wise combination with voriconazole (panel A), amphotericin B (panel B) and anidulafungin (panel C) and imipenem alone and in combination with posaconazole (panel D) against P. aeruginosa monomicrobial and Pa/Af biofilms. A 24h exposure of P. aeruginosa monomicrobial biofilm to 16µg/ml cefepime reduced CFU by 2.12 to 3.94 logs. The same concentration of cefepime reduced P. aeruginosa CFU in mixed microbial biofilm by only 0 to 0.54 logs when compared to the drug free control, suggesting that polymicrobial biofilm embedded P. aeruginosa cells were highly recalcitrant to the bactericidal activity of cefepime (p values ranged from 0.0076 to 0.0509). Similarly, evaluation of the effects of cefepime (16µg/ml) in two-drug combination with various antifungal drugs (16µg/ml) against P. aeruginosa monomicrobial (0.94 to 4.5 logs CFU reduction) and polymicrobial (0 to 0.74 logs CFU reduction) biofilms showed that the latter was substantially less susceptible to the drug combinations, except for cefepime plus amphotericin B (p values ranged from 0.0141 to 0.1370). However, a comparison of the effects of cefepime alone and in combination with various antifungal drugs against P. aeruginosa monomicrobial (p values ranged from 0.3581 to 0.4294) and Pa/Af (p values ranged from 0.0660 to 0.1702) biofilms showed no significant difference suggesting that the presence of the antifungal in combination did not affect the activity of cefepime. Panel D shows the effect of imipenem alone and in two-drug combination with posaconazole against monomicrobial and polymicrobial biofilms of P. aeruginosa and A. fumigatus. Unlike cefepime, the bactericidal activity of imipenem did not peak at 16µg/ml, but maximum effect was obtained at 64µg/ml. However, the Pa/Af biofilm (»0.5 logs CFU reduction) was significantly less susceptible to imipenem compared to its activity against monomicrobial biofilm (»1.5 logs CFU reduction) (p=0.0492). The presence of posaconazole did not affect the activity of imipenem against P. aeruginosa since its activity against polymicrobial biofilm alone and in combination with posaconazole showed no significant difference.
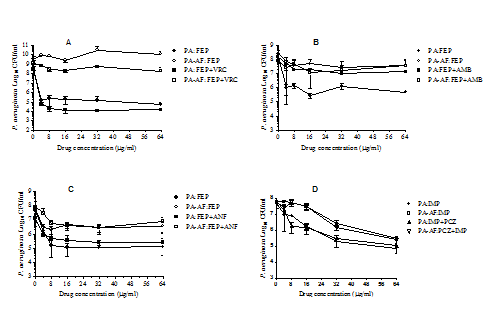
Figure 3 Effects of cefepime alone and in two-drug combinations with various antifungal drugs (Panels A-C) and imipenem alone and in combination with posaconazole (Panel D) against PA56402 monomicrobial and PA56402-AF53470 polymicrobial biofilms. The biofilms were formed in Costar 24-well cell culture plates and treated with various antimicrobial drug(s) for 24h at 35 °C in SD broth, washed and the numbers of CFU were determined by selective growth as previously described. The experiment was performed two times independently and the data were analysed by pair-wise two-tailed Student’s t-test and one-way ANOVA with Bonferroni’s post test comparison where each set of data was compared with all the other sets of data using Graphpad Prism 5.0. A p value ≤0.05 was considered to be significant. The vertical bar on each data point denotes standard deviation of two independent experiments. In addition, the experiment was repeated twice with PAO1 and AF43135 and similar results were obtained. PA, P. aeruginosa monomicrobial biofilm; PA-AF, P. aeruginosa-A. fumigatus polymicrobial biofilm; VRC, voriconazole; PCZ, posaconazole; AMB, amphotericin B; ANF, anidulafungin; FEP, cefepime; IMP, imipenem.
Figure 4 shows the effects of ciprofloxacin alone and in two-drug combination with voriconazole (panel A), posaconazole (panel B), amphotericin B (panel C) and anidulafungin (panel D) against P. aeruginosa monomicrobial and Pa/Af polymicrobial biofilms. A 24h exposure of P. aeruginosa monomicrobial biofilms to 16µg/ml ciprofloxacin reduced CFU by 2.58 to 4.65 logs, whereas the same concentration of ciprofloxacin reduced P. aeruginosa CFU in Pa/Af polymicrobial biofilms by 2.82 to 3.58 logs compared to the drug free control. This suggest that the monomicrobial and polymicrobial biofilm-embedded P. aeruginosa cells were almost equally susceptible (p values ranged from 0.1095 to 0.4339) to the bactericidal activity of ciprofloxacin. Similarly, evaluation of ciprofloxacin (16µg/ml) in two-drug combination with various antifungal drugs (16µg/ml) against P. aeruginosa monomicrobial (2.33 to 5.09 logs CFU reduction) and Pa/Af (2.94 to 4.08 logs CFU reduction) biofilms showed that they were essentially equally susceptible (p values ranged from 0.1215 to 0.4314) to the drug combinations. In addition, a comparison of the effects of ciprofloxacin alone and in two-drug combination with various antifungal drugs against P. aeruginosa monomicrobial (p values ranged from 0.1784 to 0.5835) and P. aeruginosa-A. fumigatus polymicrobial (p values ranged from 0.2150 to 0.912) biofilms showed no significant difference suggesting that the presence of the antifungal drug in the combination did not affect the activity of ciprofloxacin. Thus, the effects of ciprofloxacin alone and in combination with the antifungal drugs against P. aeruginosa monomicrobial and Pa/Af biofilms were markedly different than those seen with cefepime and imipenem alone and in combination with various antifungals. While the Pa/Af biofilm showed significantly lower susceptibility to cefepime and cefepime plus the antifungal drug combinations, both monomicrobial and Pa/Af biofilms were highly susceptible to ciprofloxacin alone and in two-drug combination with either voriconazole, posaconazole, amphotericin B and anidulafungin. The in vitro activity of the antifungal or the antibacterial drug was not affected in the presence of combination therapy, demonstrating that it is unlikely that there was any antagonistic interaction between the antibacterial and the antifungal agent.
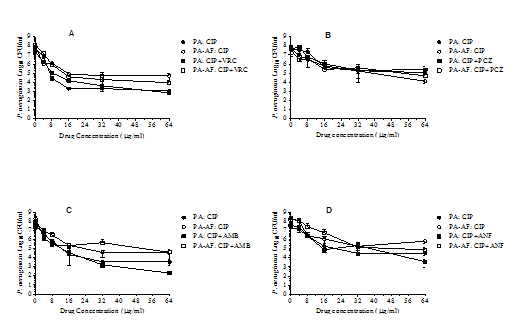
Figure 4 Effects of ciprofloxacin alone and in two-drug combinations with various antifungal drugs against PA56402 monomicrobial and PA56402-AF53470 polymicrobial biofilms. Each experiment was performed twice with PA56402 and AF53470 and once with the laboratory strains PA27853 and AF36607 (data not shown). The data were analysed by pair-wise two-tailed Student’s t-test and one-way ANOVA with Bonferroni’s post test comparison where each set of data was compared with all the other sets of data using Graphpad Prism 5.0, and a p value ≤0.05 was considered to be significant. The vertical bar on each data point denotes standard deviation of two independent experiments. Results obtained for PA27853 and AF36607 were very similar those shown in Figure 4. PA, P. aeruginosa monomicrobial biofilm; PA-AF, P. aeruginosa-A. fumigatus polymicrobial biofilm; VRC, voriconazole; PCZ, posaconazole; AMB, amphotericin B; ANF, anidulafungin.; CIP, ciprofloxacin.
Effects of antimicrobial drugs on planktonic cells: Figure 5 shows the bactericidal and fungicidal activities of various antimicrobial drugs individually and in two-drug combinations against monomicrobial and polymicrobial planktonic cultures of Pa/Af. Panel A shows the results of the CFU assay of the initial inoculum used for the experiment. The initial inoculum was standardized to 1x106 cells/organism/ml in the monomicrobial and polymicrobial cell suspensions. The assay was performed immediately after the mixed cell suspensions were prepared. Both P. aeruginosa and A. fumigatus produced approximately 1x106 CFU/ml ± experimental errors. Therefore, any reduction in CFUs obtained after the drug treatment was not due to variation in the initial inoculum.
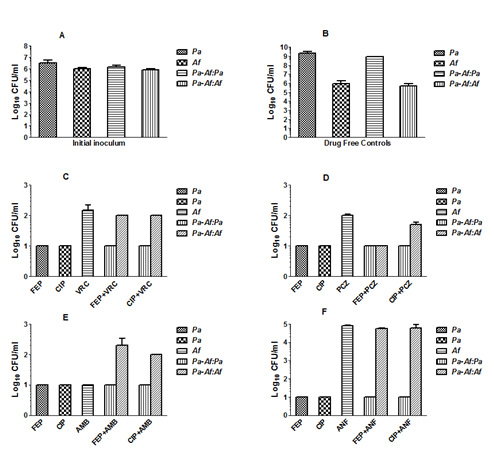
Figure 5 The microbicidal activities of various antimicrobial drugs individually and in two-drug combinations against monomicrobial and mixed microbial planktonic cultures of PA56402 and AF53470. Panel A shows the results of CFU assay for the initial inoculum. Panel B shows growth of the organism in monomicrobial and mixed microbial cultures in the absence of any antimicrobial drug (drug-free growth controls). Panels C-F shows the effects of various antimicrobial drugs alone (4 times MICs shown in Table 1) and in pair-wise combination against P. aeruginosa and A. fumigatus in monomicrobial and mixed microbial cultures. FEP, cefepime; CIP, ciprofloxacin; VRC, voriconazole; PCZ, posaconazole; AMB, amphotericin B; ANF, anidulafungin; Pa, P. aeruginosa; Af, A. fumigatus; Pa-Af:Pa, P. aeruginosa CFUs obtained in mixed cultures; Pa-Af:Af, A. fumigatus CFUs obtained in mixed cultures. Each histogram represents the mean of CFU values obtained for two independent experiments and the vertical bar denotes standard deviation. The reduction of CFUs compared to the initial inoculum ranged approximately from 4-5 logs for P. aeruginosa and 3-4 logs for A. fumigatus except for anidulafungin. All comparisons were highly significant (P≤0.001) except in the case of A. fumigatus CFU values in Panel F where the fungistatic drug anidulafungin is used. Pair-wise comparisons of the CFUs for each group were performed by Student’s t-test using Graphpad prism 5.0 for Windows (GraphPad Software, Inc., La Jolla, CA, USA). In addition, the experiment was repeated once with PAO1 and AF43135 and similar results were obtained.
Panel B shows the growth of P. aeruginosa and A. fumigatus in the absence of antimicrobial drugs (drug-free control) for 24h at 35˚C in suspension cultures. P. aeruginosa and A. fumigatus produced the same level of growth in monocultures and in mixed cultures as determined by CFU assay. These results show that any reduction of CFU demonstrated in Panels C-F is unlikely to be due to antagonistic interactions between P. aeruginosa and A. fumigatus.
A comparison of the CFU values shown in Panels C-F with the initial cell density used for the kill curve experiments (1x106 cells each/ml) showed that both cefepime and ciprofloxacin were highly effective in killing planktonic cells of P. aeruginosa (»4-5 logs reduction in CFU) in monomicrobial and polymicrobial planktonic cell suspensions. Similarly, the antifungal drugs voriconazole, posaconazole, amphotericin B and anidulafungin reduced CFU values on the average 3-4 logs except in the case of the echinocandin, anidulafungin, which is known to be a fungistatic drug against A. fumigatus and showed no significant activity. The presence of the antibacterial drug in the combination did not affect the activity of the antifungal drug and vice versa. Cefepime and ciprofloxacin provided more or less similar results in the presence of all four antifungal drugs used.
Our prior studies evaluating the formation and antimicrobial drug susceptibilities of monomicrobial and polymicrobial biofilms of A. fumigatus and P. aeruginosa indicated that A. fumigatus and P. aeruginosa are capable of co-existing in a sustainable manner in mixed microbial cultures in vitro. Microscopic examination of the monomicrobial and polymicrobial biofilms showed that P. aeruginosa produced a dense, more adherent biofilm in the presence of A. fumigatus hyphae. In part, this is due to the fungal hyphae supporting the polymicrobial biofilm production. In our recent studies using SEM, we were able to confirm our previous observations by light microscopy and revealed the presence of thicker biofilms in cocultures of Pa/Af demonstrating a firmly adhered mycelial growth as scaffolding to support the synthesis of a densely packed extracellular matrix that encases the bacterial cells. However, it is highly likely that other contributing genetic factors are in play assisting in the increased synthesis of extracellular matrix.
One of the characteristic features of the monomicrobial biofilm of A. fumigatus is the absence of hyphal branching. In the planktonic shaken or static cultures, A. fumigatus hyphae branch extensively either at the junctional point where the septum formation takes place or at the growing apical region of the hypha, producing secondary and tertiary branches forming a prolific network of mycelial growth. In contrast, the firmly adherent extensive mycelial growth in biofilm cultures seldom produces any branches and the hyphae extend by apical growth and forms bundles of parallel packed hyphal filaments fused together apparently by the extracellular.
The absence of branch formation is an intriguing observation for an organism usually thrives by producing primary and secondary branches. Branching takes place in the presence of plenty of nutrients during prolific vegetative growth conditions for the rapid expansion of fungal growth and establishes rapid infection in the case of a pathogenic organism. In contrast, during starvation or in the absence of nutrients the fungus resorts to have fewer branching, non- prolific growth and exists in a so-called ‘maintenance mode’ as opposed to active growth. Microbial biofilms are commonly produced under conditions of chronic or long-term infection not during active infection. Thus, under chronic infectious stage the microorganisms may be in the so-called ‘maintenance mode’ equipping them for survival. Therefore, it is not surprising that the fungal hyphae are not branching out vigorously under biofilm producing condition since it is functionally analogous to a state of chronicity.
Four commonly used antifungal drugs and three antibacterial drugs known to have excellent activity against P. aeruginosa were used in this study. We used the CFU assay for measuring the effectiveness of various antimicrobial drugs on the monomicrobial and polymicrobial biofilms of A. fumigatus and P. aeruginosa. The other techniques such as XTT,57,58 and MTT59,60 assays previously used for the evaluation of the effectiveness of antimicrobial drug treatment were unsuitable in these experiments because of the difficulty for distinguishing the contribution of the bacterial and the fungal cells of the polymicrobial biofilm in the reduction of the tetrazolium compound. On the other hand, since the CFU assay was based on selective growth of the bacterial and fungal cells after drug treatment, the results provided a more refined effect of the drugs on A. fumigatus and P. aeruginosa. The drawback of the CFU assay was that the hyphal form of A. fumigatus produced a high variability. This is primarily because of the tendency of the fungal hyphae for clumping and the fact that the presumed “one fungal colony for one hyphal element principle” does not apply and often results in under estimation of the CFU values. Additionally, since the clumping is a random phenomenon, high variability between replicates was commonly seen. To minimize the impact of possible poor replication and a possible under estimation of the fungal CFU values on the results, we repeated each experiment several times.
Our previous preliminary studies on the effects of the triazole antifungal drugs on the fungal constituent of the polymicrobial biofilm showed no significant change in its susceptibility compared to that in monomicrobial biofilm. In these studies, in addition to voriconazole and posaconazole, members of the polyene and the echinocandin families were evaluated. Results of this study not only confirmed our previous finding, but also demonstrate that the lack of differential susceptibility of A. fumigatus in polymicrobial biofilm is not limited to triazoles, but is also true for amphotericin B and anidulafungin, different classes of antifungals. There may be several reasons for the apparent lack of effect on the fungal hyphae including the quiescent nature of the hyphae in the biofilm and the pluripotent ability of the hyphae of filamentous fungi.
Among the four antifungal drugs used, posaconazole showed the best activity against monomicrobial and polymicrobial biofilms of A. fumigatus. However, in contrast to the excellent activity of posaconazole against conidia, sporelings and planktonic hyphae, the adherent mycelia were less susceptible to the fungicidal activity of posaconazole. Overall, anidulafungin showed the least fungicidal activity against the monomicrobial and polymicrobial biofilms of A. fumigatus. This is not surprising considering the fact that the echinocandins are known to be fungistatic agents against filamentous fungi such as A. fumigatus, and elicit their action on the apices of growing fungal hyphae by inhibiting the synthesis of new cell wall. Although anidulafungin inhibits the growth of the fungal hyphae, its inhibitory action precludes any fungicidal effect on the vegetative hyphae. Since mature fungal hyphae have the ability to grow and form new colonies, anidulafungin showed very little effect on the CFU’s of monomicrobial and polymicrobial biofilms of A. fumigatus. This observation also suggests that the reduction in fungal CFU may reflect fungicidal activity of the drug (not inhibition of fungal growth) since the triazoles and the polyenes are fungicidal for A. fumigatus.61 A comparison of the in vitro activities of several antifungals against A. fumigatus monomicrobial and Af/Pa biofilms showed no significant difference. Monomicrobial and mixed microbial cultures of A. fumigatus with P. aeruginosa were almost equally susceptible to the fungicidal activities of the antifungal drugs. Overall, 1-1.5 log CFU reductions were obtained after a 24h exposure to different drugs. Similarly, a comparison of the effect of the antifungal drug alone or in combination with the antibacterial drug showed similar fungicidal activities indicating that there was no in vitro drug-drug interaction affecting their activities.
In contrast to the similar susceptibilities of monomicrobial and polymicrobial biofilms of A. fumigatus to various antifungal drugs, the Pa/Af biofilm was substantially less susceptible to the antibacterial effect of cefepime and imipenem when compared to P. aeruginosa monomicrobial biofilm, although P. aeruginosa isolates were susceptible to cefepime and imipenem in planktonic cultures. This differential susceptibility of the monomicrobial and polymicrobial biofilms of P. aeruginosa to cefepime and imipenem was not seen when the biofilms were exposed to ciprofloxacin. At the highest drug concentration after a 24h exposure, both monomicrobial and polymicrobial biofilm-bound P. aeruginosa cells were killed almost to the same extent suggesting that certain combinations of drugs are more effective against biofilm-embedded cells than others.
In conclusion, using a previously developed 24-well cell culture plate biofilm model we investigated the antimicrobial activities of cefepime, imipenem and ciprofloxacin individually and in two-drug combinations with four antifungal drugs against monomicrobial and polymicrobial biofilms of three isolates of P. aeruginosa and A. fumigatus. Both A. fumigatus monomicrobial and Pa/Af biofilms were equally susceptible to the antifungal drugs with and without the antibacterial drugs. On the other hand, the P. aeruginosa monomicrobial and Pa/Af biofilms were almost equally susceptible to ciprofloxacin in the presence and absence of the antifungal drugs, whereas the Pa/Af biofilm complex was less susceptible to cefepime and imipenem than the P. aeruginosa in monomicrobial biofilm.
Overall, we describe that the susceptibility of A. fumigatus cells to the antifungal drugs remained almost the same in monomicrobial and polymicrobial biofilms. On the other hand, the susceptibility of polymicrobial biofilm-bound P. aeruginosa cells to cefepime and imipenem, but not to ciprofloxacin, was significantly reduced compared to the activity seen in monomicrobial biofilms.
The authors would like thank Dora Vager for her excellent technical assistance. Parts of the results included in this manuscript were previously presented at the Fifty-first Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, USA, 2011 (Abstract M-308).
This work was supported by Intramural Research Initiative from the Department of Medicine, Georgia Regents University, Augusta, Georgia, USA.
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.

©2015 Manavathu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.