Journal of
eISSN: 2373-437X


Short Communication Volume 10 Issue 6
1Laboratorio de Estequiometría and Microbiología Ambiental, Facultad de Estudios Superiores Zaragoza, Universidad Nacional Autónoma de México, México
2Laboratorio de Bacteriología Intestinal, Hospital Infantil de México Federico Gómez, Ciudad de México, México
3Laboratorio de Evolución Molecular and Experimental, Instituto de Ecología, Universidad Nacional Autónoma de México, México
4Departamento de Microbiología and Parasitología, Facultad de Medicina, Universidad Nacional Autónoma de México, México
5Laboratorio de Aerobiología, Instituto de Ciencias de la Atmósfera y Cambio Climático, Universidad Nacional Autónoma de México, México
Correspondence: Irma Rosas-Pérez, Instituto de Ciencias de la Atmósfera y Cambio Climático, Universidad Nacional Autónoma de México, Circuito de la Investigación Científica S/N, Ciudad Universitaria, Ciudad de México, México, Tel (0155) 5622 4800
Received: November 21, 2022 | Published: November 28, 2022
Citation: Valdivia-Anistro J, Cruz-Cordova A, Souza V, et al. Diversity of cultivated methylotrophs from the extremely oligotrophic system in the Cuatro Cienegas Basin, Mexico: An unexplored ecological guild. J Microbiol Exp. 2022;10(6):208-214. DOI: 10.15406/jmen.2022.10.00375
The simplest form of heterotrophy in the carbon cycle is to metabolize C1 compounds, this is a widely spread strategy that includes genus in different phyla inhabiting diverse environments that seem to have acquired the methanol dehydrogenase by horizontal gene transfer (HGT). The objective of this study was to isolate and explore the diversity of the ecological guild of methylotrophs in the water and riparian vegetation of the Churince system in the Cuatro Cienegas Basin (CCB), Coahuila, Mexico. Methylotrophy was verified by polymerase chain reaction (PCR) amplification of the mxaF gene that encodes the α-subunit of the enzyme methanol dehydrogenase (MDH), while phylogenetic affiliations were assigned following 16S rRNA phylogenetic analyses. Among the isolated strains we observed a phylogenetic association with a common species of Methylobacterium (M. radiotolerans). In addition, other methylotrophs were isolated, like Methylorubrum aminovorans, Methylorubrum extorquems and Methylophilus methylotrophus. Interestingly, we also isolated other strains able to grow in methanol and mxaF+, their 16S rRNA identified them as Jiella, Pseudomonas, Rhizobium, Serratia and Stenotrophomonas. This study addresses, for the first time, the diversity of cultivated methylotrophic bacteria within CCB and inserts this knowledge in the context of a total inventory of the microbiota in the site.
Keywords: methylotrophy, Methylobacterium, Cuatro Cienegas Basin, oligotrophic environment
Methylotrophy is the ability to metabolize reduced single carbon (C1) compounds (i.e., methanol and methylamine), it encompasses diverse metabolic capabilities that represent an ecological guild per se in the Carbon cycle.1,2 Moreover, methylotrophy represents a phenomenon more widespread in the phylogenetic tree and ecological versatile than initially assumed.3,4 Such genetic and biochemical diversity may explain the ecological success of methylotrophic bacteria, which can represent dominant microbial populations in specific environments such as plant leaves.5,6 Within this ecological guild, the genus Methylobacterium is one of the best understood. Species within this group consist of aerobic, facultative α-proteobacteria capable of growing on C1 compounds such as methanol, formate and formaldehyde as well as a range of multi-carbon complexes.2,7 In 2011, the genome of the facultative methylobacteria Methylobacterium extorquens was analyzed.8 The metabolic network of the bacterium reconstructed contains 1139 reactions and 977 metabolites. The authors observed that the core metabolism has a highly unusual topology, in which the unique enzymes that catalyze the key steps of C1 assimilation are tightly connected by several, large metabolic cycles. This observation suggests that in nature a strong pressure of selection must exist to maintain the methylotrophic capability. Nevertheless, the metabolic network is highly versatile around a flexible backbone of central reactions that allows rapid switching to multi-carbon sources. As a result, Methylobacterium are ubiquitous in nature.9-11
Members of the genus Methylobacterium are one of the major inhabitants of the phyllosphere12-14 which is recognized as an environment with little protection from extremes of temperature, UV radiation and desiccation.15-17 In plants that live under dry conditions, Methylobacterium can help in their growth with the production of phytohormones and in the survival given osmoprotectants from desiccation.18
The Cuatro Cienegas Basin (hereafter CCB) is an extremely diverse and oligotrophic oasis in the Chihuahuan desert in the Mexican state of Coahuila.19-21 Elemental composition at the CCB is anomalous with regards to other similar environments (N:P ratio can range from extremely low P (157:1) to very low N (1.8:1)).22 This stoichiometric imbalance causes changes of bacterial communities composition.21,22 These unusual and extreme stoichiometric ratios at the CCB generate nutrimental stresses along with high UV light and high desiccation rates in some shallow ponds such as Churince lagoon. We believe that such extreme conditions may dictate population dynamics perhaps more akin to leaves in the case of methylotrophs than to normal wetlands. In this study, we isolate and explore the diversity of C1 heterotrophs from both environments: the highly oligotrophic water column (<1.0 μmol PO43-, high concentrations of sulfate and carbonate ions),23 and the phyllosphere within surrounding riparian vegetation in the Churince system.
Site description
The Churince system is located to the west of the arid basin of the CCB, Mexico. It consists of freshwater spring “Manantial” (M) (26°50.421’N, 102°08.053’W) which connects to an intermediate lagoon, the “Laguna Intermedia” (LI) (26°50.913’N, 102°08.574’W), via a small river. The aquatic system terminates in a shallow desiccation lagoon, the “Laguna Grande” (LG) (26°50.830’N, 102°09.335’W), that went practically extinct due to the over-exploitation of the aquifer Figure 1.

Figure 1 Google earth image of the Churince system in the Cuatro Cienegas Basin, Mexico. a, Laguna Grande; b, Laguna Intermedia; c, Manantial; d, plant of the genus Asclepias.
The dissolved ionic composition of the water body is characterized by high concentrations of sulphates (SO42-) (2700-3100 mg/L), calcium (Ca2+) (560-620 mg/L) and carbonates (CO32-) (121-125 mg/L). Typically, the ratio of C:N:P in the water column is 1000:100:1 indicating extreme oligotrophy.20 Moreover, the soil within the basin contains high concentrations of sulphates, preventing total colonization by plants.24 This is, however, except for members of the genus Asclepias (Figure 1d), which grow adjacent to various water bodies within the system.
Sample collection, strain isolation and culture conditions
Surface water and riparian vegetation were sampled from three sites (M, LI and LG). Fifteen surface water samples (10-30 cm in depth) were retrieved in sterile tubes and processed within one hour of collection, and 12 leaf samples were collected adjacent to the sites where water samples were retrieved.
Methylotrophic bacteria were isolated using a minimal ammonium mineral salt agar medium (AMS per liter: 0.7 g K2HPO4, 0.54 g KH2PO4, 1.0 g MgSO4•7H2O, 0.2 g CaCl2•2H2O, 4.0 mg FeSO4•7H2O, 0.5 g NH4Cl, 100 μg ZnSO4•7H2O, 30 μg MnCl2•4H2O, 300 μg H3BO3, 200 μg CoCl2•6H2O, 10 μg CuCl2•2H2O, 20 μg NiCl2•6H2O, 60 μg Na2MoO4•2H2O, 100 mg cycloheximide and 15 g agar-agar; supplemented with 0.5% methanol like only carbon source).25 One hundred microliters of each water sample were spread directly onto methanol agar plates. Leaf samples were processed by the imprinting technique;26 however, intense fungal growth inhibited the development of bacterial colonies. To prevent this, 1.0 g of leaf sample was transferred into a tube containing washing solution (9.0 mL of 0.85% NaCl solution with 2.5 μL of 0.025% Tween solution). The sample was subsequently sonicated for 20 min in an ultrasonic cleaning bath (Branson 5210) to dislodge bacteria from each leaf. Serial dilutions were performed to a concentration representing 10-3 relative to original samples and 100 μL of diluted samples were spread onto ASM agar. Each sample was spread by triplicate.
Plates were initially incubated at 28°C for seven days. During this period, however, in some samples the growth of yellow- and white-pigmented colonies vastly outnumbered the expected pink-pigmented colonies, typically associated with Methylobacterium.7 Consequently, the incubation period was extended for a further eight days, thus permitting the formation of pink-pigmented colonies. Single strain cultivation was followed to obtain single colony pure cultivars that were classified by color. The purified cultures were maintained on ASM liquid medium with glycerol (20% v/v) at -70°C, until their use.
Genomic DNA extraction and amplification of mxaF gene
DNA extractions were performed using the protocol described by Hassan.27 The mxaF gene, which catalyzes the oxidation of methanol to formaldehyde in methylotrophic bacteria and has previously been employed for the authentication of genus Methylobacterium, was used in PCR amplification.9,12,28 The mxaF gene (550bp) was amplified using the forward primer mxaF (5'-GCG GCA CCA ACT GGG GCT GGT-3') and reverse primer mxaR (5'-GGG CAG CAT GAA GGG CTC CC-3'). Each PCR reaction was performed using positive (Methylobacterium extorquens ATCC14718) and negative (ultra-high water) controls. The conditions of PCR amplification were as follows: 94°C for 1 min for the initial denaturation, 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 30 s, an extension of 1 min at 72°C and a final extension of 1 min at 72°C (Gene Amp, PCR System 9700). The presence and size of PCR products were subsequently confirmed by 1.5% agarose gel electrophoresis.
Amplification of the 16S rRNA gene and ribotyping analysis
The 16S rRNA gene was amplified using the universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1392R (5’-GGTTACCTTGTTACGACTT-3’). The PCR amplification was according to the following program: 94°C for 1 min of initial denaturation, 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 30 s, an extension of 1:30 min at 72°C and a final extension of 5 min at 72°C (Gene Amp, PCR System 9700). The presence and size of amplicons were confirmed by 1.5% agarose gel electrophoresis.
The restriction enzyme selection was fulfilled with the NEBcutter V2.0 program,29 using the 16S rRNA available sequences from the Methylobacterium type strains from the GenBank database (NCBI). The enzymes selected to generate ribotypes for this genus were KpnI, XhoI and StuI (New England BioLabs ® Inc.) (Figure 2). The PCR products of 16S rRNA gene were restricted at 30°C for 2 hrs. Each digestion was performed using the 16S rRNA PCR product of M. extorquens ATCC14718 like positive control. The restriction fragments were separated by electrophoresis in 1.5% agarose gel.
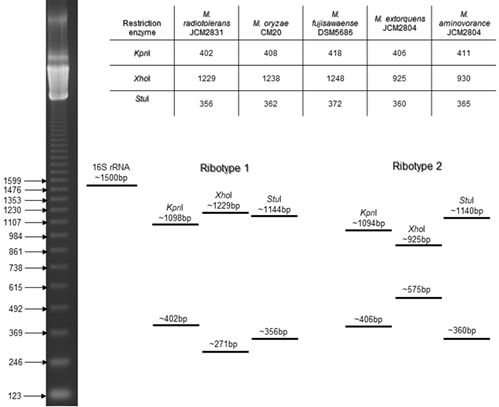
Figure 2 Restriction enzymes selected with the in silico tool NEBcutter V2.0 for ribotyping. Cutting sites of the restriction enzymes in the 16S rRNA sequences of Methylobacterium strains from the GenBank database (NCBI), and the expected ribotypes: ribotype 1, generated with the 16S rRNA sequence of Methylobacterium radiotolerans JCM2831 and ribotype 2, generated with the 16S rRNA sequence of M. extorquens JCM2804.
Phylogenetic analysis
For phylogenetic analysis, the 16S rRNA gene was amplified using DNA extracted with the QIAmp® DNA Mini DNA Kit (USA), according to the manufacturer’s instructions. The PCR conditions were those previously described. The presence and size of amplicons were confirmed by 1.5% agarose gel electrophoresis. All PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) (USA) for sequencing (High Throughput Genomics Center University of Washington, USA).
The resulting 16S rRNA gene sequences were compared with the available sequences from the GenBank database (NCBI) using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences were aligned and designated according to their phylogenetic affiliation using CLUSTAL W.30 Phylogenetic trees were constructed using the maximum-likelihood method in the program MEGA X31, with a bootstrap of 1000 replicates.
Quantification and isolation of different pigmented bacterial colonies
In the minimal ammonium mineral salt agar medium, in addition to the expected formation of pink-pigmented colonies (PP), the formation of white- (WP) and yellow-pigmented (YP) colonies occurred within the first seven days of incubation. These white and yellow colonies represented the dominant form isolates relative to pink-pigmented colonies. While pink-pigmented colonies were identified until 15 days of incubation. Yellow-pigmented colonies were isolated from six water samples, whilst both pink- and white-pigmented colonies were isolated from the remaining samples. The numbers of colony-forming units (CFU) were calculated per milliliter (mL) and gram (g), respectively (Figure 3). The total number of isolates derived from water and leaf samples were 171 (PP= 73; WP= 80; YP= 18) and 315 (PP= 100; WP= 215), respectively.

Figure 3 Colony-forming units (CFU) of pigmented methylotrophic bacteria isolated from (a) water and (b) phyllosphere samples in the Churince system. White- pigmented (WP) colonies are represented by the white and hatching bars; yellow-pigmented (YP) colonies by black bars and pink-pigmented (PP) colonies by the grey bars. Water samples: 1 to 5: “Laguna Grande” (LG); 6 to 14: “Laguna Intermedia” (LI); 15: “Manantial” (M). Phyllosphere samples: 1 and 2: “Laguna Grande” (LG); 3 to 11: “Laguna Intermedia” (LI); 12: “Manantial” (M).
Selection and verification of Methylobacterium isolates
Of the total number of 486 strains, 130 pink-, 150 white- and 18 yellow-pigmented colonies were selected for the PCR amplification of the mxaF gene, a marker for the use of methanol as a source of carbon and energy among isolates. Only 44% (57/130) of pink-pigmented and 32% (53/168) isolates of different pigmentation amplified the expected PCR-product of 550 bp (MDH+) (Figure 4).
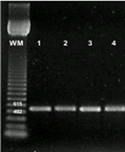
Figure 4 PCR amplification of mxaF gene from methylotrophic bacteria. Lane WM, Weight marker: 123 bp DNA Ladder, Invitrogen. Lane 1, Methylobacterium extorquens ATCC14718; 2. pink-pigmented; 3, white-pigmented; 4, yellow-pigmented colonies.
Furthermore, all the MDH+ isolates were subject to restriction digestion of 16S rRNA gene using three endonucleases to generate ribotypes for genus Methylobacterium (Figure 5). According to the simulations generated with the NEBcutter V2.0 program, 35 of the pink-pigmented isolates produced the most common restriction pattern (Ribotype 1) and six isolates showed sizes variations in the fragments restricted with the endonuclease XhoI (Ribotype 2); the remaining isolates did not have digestion fragments with the endonuclease XhoI (Ribotype 3). As expected, among the isolates with different pigmentation, just five white- and four yellow-pigmented colonies yielded the common restriction pattern expected for Methylobacterium.

Figure 5 Ribotyping analysis of methylotrophic bacteria from Churince system. From every ribotype: first line, restriction fragment obtained with KpnI; second line, restriction fragment obtained with XhoI; third line, restriction fragment obtained with StuI. A: aquatic- and F: phyllosphere-isolate, respectively. Isolates A85 and F71, are white-pigmented. Isolate A72, is yellow-pigmented. Isolates A31, A43, A57, F13, F22, F40, F68 and F99, are pink-pigmented. Lane WM, Weight marker: 123 bp DNA Ladder, Invitrogen.
For 16S rRNA sequencing we selected the isolates that were MDH+ with restriction fragments, these included 41 pink-pigmented colonies together with the nine isolates of different pigmentation.
16S rDNA phylogenetic analysis
Partial 16S rRNA gene sequences with an average length of 640 nucleotides (minimum, 580 nucleotides; maximum, 700 nucleotides) were analyzed and compared with sequences in the GenBank database. Only nearest neighbors sequences were selected. To confirm the phylogenetic position of each sequence, 18 isolates were selected for complete 16S rRNA gene sequencing (average length of 1363 nucleotides). Phylogenetic analyses revealed that 86% of these isolates were affiliated with the α-proteobacteria subdivision, 4% to β- and 10% to γ-proteobacteria subdivisions.
Of the 22 isolates retrieved from water samples, 15 were affiliated with the α-proteobacteria (Figure 6). Within this subdivision, members of the genus Methylobacterium were identified. These included 12 isolates related to strains of M. radiotolerans (98-100% identity) and one strain of Methylobacterium sp. F73 (99.78% identity). In addition, two isolates were related to Methylorubrum aminovorans CCM4612T (99.75% identity; formerly Methylobacterium aminovorans).
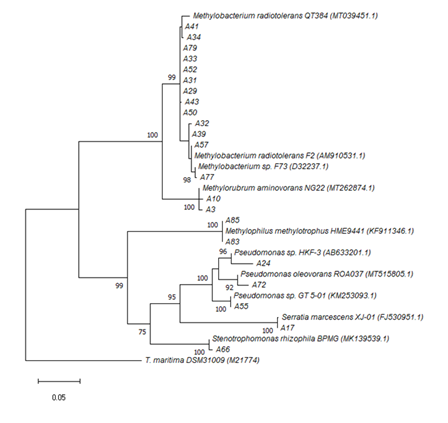
Figure 6 Phylogenetic tree obtained by the maximum-likelihood method showing the relationship between the 16S rRNA gene sequences form isolates of the Churince aquatic system. Samples were retrieved from sites: “Laguna Grande” (LG); “Laguna Intermedia” (LI); “Manantial” (M). Bootstrap values above 70% are shown. The scale bar represents the number of changes per nucleotide position. Accession numbers of environmental sequences are in parenthesis. Thermotoga maritima DSM31009 represents the out group.
Among leaf samples, 20 isolates were identified that shared high sequence similarity with two strains of M. radiotolerans: strain F2 (four isolates; 99.78 to 100% sequence identity) and strain QT384 (16 isolates; 99.84 to 100% sequence identity). Also, two isolates were affiliated with the strain Methylobacterium sp. D2P087 (99.69% sequence identity), which forms part of bacterial the community of leaves. The sequence F30 shared 99.78% identity with the strain Methylorubrum extorquens. This is the type species of the new genus proposed to reclassify several species of Methylobacterium.32 (Figure 7)
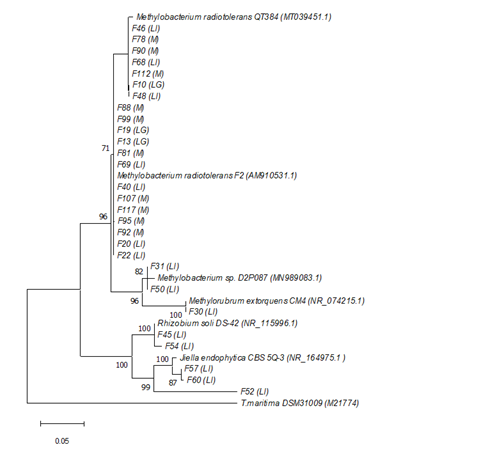
Figure 7 Phylogenetic tree obtained by the maximum-likelihood method showing the relationship between the 16S rRNA gene sequences from phyllosphere isolates of the Churince system. Sampled were retrieved from sites: “Laguna Grande” (LG); “Laguna Intermedia” (LI); “Manantial” (M). Bootstrap values above 70% are shown. The scale bar represents the number of changes per nucleotide position. Accession numbers of environmental sequences are in parenthesis. Thermotoga maritima DSM31009 represents the outgroup.
Seven isolates were identified as members of the Proteobacteria, among water samples. Of these, the sequences A83 and A85 shared 100 and 99.57% similarity, respectively, with the methanol-utilizing bacterium Methylophilus methylotrophus (β-proteobacteria).33 In total, five isolates were affiliated with the γ-proteobacteria subdivision. Among these, isolate A17 shared 99.79% sequence identity with Serratia marcescens XJ-01; three isolates with the Pseudomonas genus: P. sp. HKF-3 (97.13% identity; isolate A24), P. sp. GT 5-01 (99.86% identity; isolate A55) and P. oleovorans ROA037 (98.64% identity; isolate A72). An additional, the sequence A66 shared 99.57% identity with Stenotrophomonas rhizophila FB206, isolated from the evergreen conifer Pseudotsuga menziesii. (Figure 6)
Regarding leaf samples, five additional isolates were affiliated with the α-proteobacteria subdivision. Of these, two isolates were related to Rhizobium soli strain DS-42 (99-100% sequence identity) from soil of the Dokdo island (Korea). The other three sequences were related to the strain Jiella endophytica CBS 5Q-3, with a minimum of 90.89% (F57) and a maximum of 99.53% (F57) sequence identity.(Figure 7)
The C1 ecological guild of methylotrophic bacteria has been isolated from a wide variety of aquatic and terrestrial ecosystems pointing towards a broad environmental distribution.34,35 However, in CCB, despite the ample microbial sampling,21,36,37 this is the first study where we explicitly investigate the diversity of this ecological guild in the Churince system. Herein, we observed that in addition to the expected members of the Methylobacterium group, an unexpected diversity of potentially methylotrophic bacteria and other bacteria were detected among our isolates. This may therefore suggest that the diversity of methylotrophic bacteria is greater within Churince than previously acknowledged elsewhere.
The isolation and culture of methylotrophs is the first step to understand their ecological and biotechnological potential.11,38,39 But members of the Methylobacterium group grow slowly in pure culture. The suggested incubation period, for the growth of ‘signature’ pink-pigmented colonies, is seven days at 28°C.40,41 However, in Methylobacterium from the potable water system, the incubation period was not enough for the growth of pink-pigmented colonies. In this case, the high concentrations of chlorine, within the system, were attributed to the need of increasing incubation time42,43 but the same occurred for Methylobacterium in rhizosphere, where the plates were also incubated for 14 days.18 These varied conditions suggest that most Methylobacterium are slow growers in culture and that the initial seven-day period is an underestimation, as was the case of our samples from either the phyllosphere or the water column of Churince where white- and yellow-pigmented colonies grew faster suggesting that they are better adapted to the selective media than those that form pink-pigmented colonies.
The development of typically pink-pigmented colonies among Methylobacterium, is associated with the production of an oxo-carotenoid substance, sharing a similar structure to rhodoxanthin.11,44 Essentially, these pink pigments serve as photoprotectors, critically reducing the damage generated by solar radiation.45 Given the shallow water column46 the extreme oligotrophy and direct exposure of the phyllosphere to intense UV radiation as well as desiccation,16,48 the dominance of pink-pigmented colonies were expected in the Churince system; however, the white- and yellow-pigmentation also offer protection against UV stress.48,49 In addition, in a study by Zenoff and colaborators50 in other high UV irradiated wetlands methylotrophs with different pigmented colonies where isolated in the water column.
Independently from the pigment that is typical of Methylobacterium, the most important feature of the guild of aerobic methylotrophic bacteria is the presence of the mxaF gene that encodes for the active site of the enzyme MDH (the locus of the methanol oxidation to formaldehyde).9,5,52 Consequently, amplification of this gene also serves as an effective marker for the authentication of the genus Methylobacterium along with the restriction enzyme pattern of the gene 16S rRNA.18,28 In preliminary studies, this gene was mainly identified among organisms grouping within the α-, β- and γ-proteobacteria.51 In our study, it was not possible to amplify the mxaF gene among most of the white and yellow colonies, although methanol represented the only carbon source for growth in the media. This may indicate the use of alternative enzymes for methanol oxidation among unidentified colonies, as described in other methylotrophic bacteria.4,53
Not surprising, the dominant specie in both environments was strongly related to the cosmopolitan M. radiotolerans, that has been found in all types of environments, including some extreme sites with high UV and gamma radiation54,55 as well others with high concentrations of heavy metals.56,57 Furthermore, M. radiotolerans is a plant-growth promoting bacteria with the ability to fix nitrogen, and with antimicrobial and antioxidant properties.11,58
Within the CCB methylotrophic bacteria falling outside the Methylobacterium group, members of Methylorubrum (α-proteobacteria) and Methylophilus (β-proteobacteria) were confirmed. In 2018 was proposed the taxonomic reassignment of 11 species of Methylobacterium into the new genus Methylorubrum.32 This reclassification was supported by the divergence in genetic and phenotypic traits. However, the genomic analysis of more than a thousand type-strains of the class α-proteobacteria showed that these 11 species should continue to be considered within Methylobacterium.59 Methylophilus methylotrophus is an obligate methylotroph which uses methanol as the only source of carbon and energy.33 This bacterium is commonly isolated from aquatic environments and is used for L-lysine production.60,61 Other interesting isolates, which were not directly affiliated with this metabolic group, included strains of Pseudomonas. Previous studies have demonstrated that this versatile genus can use methanol as a primary carbon source,62,63 as well high concentrations of formaldehyde to obtain energy,64 but it has never been classified among the methylotrophic bacteria. These results suggest that this metabolic guild has been largely underexplored in many habitats, including CCB were Serratia and Stenotrophomonas, seem to grow in the proper media as well present the MDH+ locus, suggesting that these aquatic microorganisms are able to metabolize methanol. This is also clearly the case concerning to phyllosphere isolates of Rhizobium and Jiella. Given that only isolates carrying the mxaF gene were subject to additional phylogenetic analysis, further exploration to confirm the identity of these unusual colonies is required. In addition, the Churince system showed a large diversity as well as a unique community composition in each site where the only common feature is that actinobacteria are the most predominant phyla in all environments.65,66 A closer look to those studies we will determine the relative abundance of potential methylotrophic bacteria, this will be complemented with metagenomics and transcriptomics. Nevertheless, despite all the technology, future “classic microbiology” studies must search and characterize a larger range of isolates that are capable of growth in minimal media with C1 compounds as a carbon source to have a better picture of this niche.
Thus, our results show that many methylotrophic bacteria inhabit the water and riparian vegetation of the Churince system in the CCB. However, there was an evident dominance of the genus Methylobacterium across all samples. Further insights are needed to determine the genetic diversity and the trophic interactions among these methylotrophic bacteria to understand the carbon cycle in this extreme oligotrophic ecosystem.
We would like to express our thanks to M. E. Salinas-Cortes and L. Martínez-Romero for technical assistance on methods. We dedicate this article to the memory of Jesús Caballero-Mellado, CCG (UNAM).
The authors declare that there is no conflict of interest.
All authors listed have made a substantial and intellectual contribution to the work.
This study was partially funded by a grant 50507 from CONACyT and from Alianza WWF-Fundación Carlos Slim (VS). This study was supported by the project PAPIME- PE204517, DGAPA, UNAM (JVA).
Not applicable.

©2022 Valdivia-Anistro, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.