Journal of
eISSN: 2373-437X


Research Article Volume 12 Issue 1
1Carrera de Biología, Facultad de Estudios Superiores de Zaragoza, Universidad Nacional Autónoma de México, Mexico
2Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad Nacional Autónoma de México, México
3Departamento de Ingeniería Química, Facultad de Química, Universidad Nacional Autónoma de México, México
4Instituto de Ingeniería, Coordinación de Ingeniería Ambiental, Universidad Nacional Autónoma de México, Mexico
5Laboratorio de Investigación en Procesos Avanzados de Tratamiento de Aguas, Unidad Académica Juriquilla, Instituto de Ingeniería, Universidad Nacional Autónoma de México, México
Correspondence: MA Alvarez-Amparán, Departamento de Ingeniería Química, Facultad de Química, Universidad Nacional Autónoma de México, 04510 Ciudad de Mexico, México
Received: February 10, 2024 | Published: February 21, 2024
Citation: Castillo-Villanueva E, Alvarez-Amparán MA, Valdivia-Anistro J, et al. Developing lateral-flow devices for the fast and cheap detection of SARS-cov-2 in wastewater: a potential tool to monitoring local virus outbreaks by wastewater based epidemiology. J Microbial Exp. 2024;12(1):16-22. DOI: 10.15406/jmen.2024.12.00410
The SARS-CoV-2 virus generates severe respiratory tract complications such as pneumonia and bronchitis and mild symptoms such as common colds or asymptomatic conditions. The SARS-CoV-2 presence in human feces and in treated/untreated wastewater suggests a transmission way that could generate local outbreaks, in addition to other type of diseases or disorders. Based on the above, in this work it was proposed the assembly of a lateral flow device (LFD) to determine the SARS-CoV-2 presence in wastewater samples. In the LFD a wastewater sample capillary flowed through four membranes: sample zone, conjugate delivery zone, reaction zone and the reactive adsorption zone. The virus amplification was achieved by the novel reverse transcription loop-mediated isothermal amplification (RT-LAMP) at the sampling point. The membranes preconditioning processes and the use of membranes with 5-20 nm porous size increased the capillary flow rate and it was promoted the interaction of the gen of SARS-CoV-2 with the capture agents in the reactive adsorption zone. Additionally, the sensibility of the detection was improved using several methods for the immobilization of the capture agents on the reaction zone membrane. The RT-LAMP method combined with the assembled LFD allowed an efficient SARS-CoV-2 detection at the sampling point in a simple way, cheap and fast compared to conventional and expensive RT-PCR.
Keywords: SARS-CoV-2 detection, SARS-CoV-2 in wastewater, point-of-care devices (POC), LAMP RNA-virus amplification, wastewater-based epidemiology (WBE), lateral flow devices (LFDs)
(HCoV), Human Corona virus; (COVID-19), coronavirus-disease-2019; (WHO), World Health Organization; (LAMP), loop-mediated isothermal amplification; (LFDs), lateral flow devices
SARS-CoV-2 virus, named by the International Committee on Taxonomy of Viruses,1 is an α-coronavirus that belongs to the family of Coronaviridae. Its crown-shaped viral cover is a characteristic of this virus family.2−4 The α-and β-corona virus are recognized as human corona virus (HCoV) and are the cause of disorders and/or severe respiratory tract complications in immunocompromised people, matured people and newly born. HCoV such as SARS-CoV, MERS-CoV y SARS-CoV-2 is responsible for severe lower respiratory tract acute conditions such as pneumonia and bronchitis although mild symptoms such as common colds or asymptomatic conditions have also been observed.6−12 In March 2020 the SARS-CoV-2 virus caused the COVID-19 (coronavirus-disease-2019 acronym) which was classified as a pandemic by the World Health Organization (WHO).1,13−16 According to the Johns Hopkins University, in August 2020, 17,639,185 COVID-19 confirmed cases were recorded and resulting in 680,575 worldwide deaths.16 Based on the above data and other statistics reported by the WHO,11 it was determined a SARS-CoV-2 virus fatality rate of 5.3-8.4%18 and the reproduction number of 1.4-6.5 (estimated number of infections for each infected patient).17−19 These values are higher to pandemics or epidemics caused by virus in the past. Also, it has been reported a very high rate of SARS-CoV-2 virus transmission which is bigger than other viruses.20
To avoid SARS-CoV-2 infections and COVID-19 disease outbreaks it is necessary to detect the virus.21 The virus detection process is carried out in three main steps: 1) the concentration and extraction of the RNA of the virus, 2) the viral gene amplification and 3) the viral detection.22 The polymerase chain reaction (RT-PCR or PCR) is the most conventional technique to the amplification of genetic sequences (RNA and DNA respectively) in temperature cycles and the real time quantitative PCR (qRT-PCR) is used complementarily to the virus identification and quantification. However, this technique is expensive and requires qualified personal to the operation.23 Other popular methods to the virus identification are CRISPR-CAS12 or SHERLOCK based on CRISPR which also need professional personal for its operation.24,25 Also, serological tests can be carried out in humans to the virus identification by means of the antibodies generated into the host.26
The virus gene amplification can be achieved using isothermal amplification methods (for the RNA or DNA viral genomes) together with another technique for the identification of the virus (qualitatively or quantitative). The loop-mediated isothermal amplification (LAMP) is a novel methodology for the amplification of genetic sequences in a specific, easy and fast way. LAMP is a low-cost technique compared with conventional methodologies. Also, LAMP technique can be carried out at the sampling point without the necessity of expensive material or tools.27,28
The SARS-CoV-2 transmission in humans compared to other HCoV types is by droplets generated by breathing, sneezing, coughing, sputum, as well as by direct contact with infected people or surfaces that contain the virus.26−29 Several studies have shown the SARS-CoV-2 presence in feces and urine of infected people, the presence of the virus has also been determined in wastewater, groundwater, drinking water, seawater, soil, sediment, and sludge; the presence of the SARS-CoV-2 virus and its viral RNA in human feces has been reported in recent studies.30−40 The presence of virus in water bodies could promote the transmission of diseases and local infections. Even the WHO (World Health Organization) have stablished procedures to the water treatment to the eradication of known virus, their persistence time could be enough to reach other organisms, mutate and/or change their characteristics.41
Virus monitoring studies in wastewater are designed primarily to detect enteric viruses without viral envelope, which are responsible for infections of the gastrointestinal system and are also associated with possible upper respiratory tract infections.41−44 Viruses with viral envelopes (such as HCoV) have not been widely monitored in wastewater. However, some types of HCoV have been detected in human feces,31,32 which suggests a possible route of fecal-oral transmission. Therefore, it is important to develop and standardize methods for the detection of HCoV in “wastewater”. Some studies have shown the presence of HCoV in wastewater and treated water.45−48 In the 2004 SARS (a type of HCoV) outbreak in Beijing, China, some publications reported the presence of the virus in treated or disinfected water from hospitals treating infected patients49 and a SARS outbreak was also associated in a residential complex due to the presence of this virus in the water system.50
Water is a vehicle for the dispersion of pathogens, promoting infectious outbreaks. Through RT-qPCR analysis, concentrations of up to 106 virus copies/liter were determined in treated or untreated water in Italy, China, Israel, Turkey, Spain, the Netherlands, the United States, France, and Australia.51−57 However, due to the few studies carried out with viral envelope viruses, the analysis method for the detection of the SARS-CoV-2 virus has not been standardized. In this sense, the methods of extraction and concentration of the virus should be improved taking as a precedent the procedures reported for other types of viruses.42−44 Currently, the RT-qPCR technique is the most widely used to validate and quantify the presence of the virus; however, it is necessary to establish the differences and similarities between the preparation of environmental samples and clinical diagnostic samples.58
Detection of various agents, including nucleic acid sequences, can also be performed by point-of-care (POC) detection devices, which are specifically designed for the detection of target analytes. Its main characteristic is the portability and ease of being carried out at the sampling site or without the need to purchase and use expensive and complex equipment for detection. These devices have been used for early disease diagnosis, food safety monitoring, and environmental contamination, and allow the presence of target analytes to be determined by simple visual inspection by colorimetry, mainly using Au nanoparticles.59−65 Detection is based on 1) pretreatment and 2) determination of target analytes (nucleic acids, proteins, ions, or chemical molecules). However, sample pretreatment and DNA/RNA extraction methods need to be further developed for greater efficiency. POC devices specifically designed for the detection of target analytes, without considering the DNA/RNA extraction itself, are more efficient and are known as lateral flow devices (LFDs). LFDs are sensitive, specific, rapid, portable, inexpensive, easy to use, biocompatible, and contain capture agents to detect target analytes.66 The detection is carried out by means of the capillary flow of the sample through different membranes, whose porous structure and surface chemical characteristics allow the detection of target analytes, through the adsorption of the target analyte with capture agents on the surface of the membranes. Physicochemical properties, such as pore size, surface area, and hydrophobicity/hydrophilicity, affect capillary forces and determine capillary flow velocity and adsorption force of target analytes. The selection of membranes must be made based on the adsorption capacity and balance, as well as their surface properties. The presence of superficial polar groups, such as hydroxyl and carboxylic groups is highly desired due to the strong interaction with the peptide bonds of proteins, which are used as capture agents, in addition to allowing specific binding with the protein. Additionally, materials must allow for adequate capillary flow time to prevent membrane wettability from significantly decreasing the adsorption capacity of capture agents. Cellulose, cotton, fabric, and fiberglass-based membranes are commonly used because they meet the mentioned characteristics. However, cellulose-based composites are currently beginning to be used with specifically designed fabrics, polyester, fibers or composites. The use of different blocking, dragging and conditioning solutions to provide the surface with hydroxyl and carboxyl groups has also been reported.67−69
Information collected through nucleic acid detection can be used for epidemiological monitoring based on wastewater studies (WBEs). WBE monitoring is commonly performed to track drugs, pesticides, personal care products, persistent organic pollutants, pathogens, among others.70−74 This approach could be successfully used to monitor the presence of the SARS-CoV-2 virus in wastewater and take corresponding sanitary actions. Ahmed et al., 2020 determined, by means of numerical simulations using the Monte Carlo method, the number of people infected as a function of the concentration of the SARS-CoV-2 virus (determined by RT-qPCR) in wastewater samples. In this study, 171-1090 infected were estimated, which was consistent with the clinical observations carried out in Australia, although they recommended further validation of the methodology to obtain more precise and conclusive results.75 It has been proposed that WBE monitoring could be used to monitor the presence of the SARS-CoV-2 virus in wastewater, which would make it possible to identify the geographic distribution of the excreted virus within communities.76 In addition, the evolution of the virus would be monitored even when its presence is not evident through clinical diagnosis due to the asymptomatic nature of patients and super saturation of clinical cases.77−80 Also, the efficiency of public health measures such as social distancing, isolation or emergency closures could be determined as interventions to prevent the spread of COVID-19.81 Nemudryi et al., 2020 reported that some states in the United States of America have adopted wastewater surveillance to determine the phylogeny of SARS-CoV-2 virus strains and monitor the efficacy of public health interventions,82 similarly studies have been proposed in the Netherlands, France and Australia.83,51
This work proposes the new assemble of lateral flow devices coupled with RT-LAMP molecular reactions to obtain a fast and cheap method for the detection of genetic fragments of SARS-CoV-2 in wastewater treatment plants and local water systems.
Materials
FTIC, Streptavidine, Biotina, anti-FTIC, anti-biotin, Bovine serum albumin (BSA), phosphates buffer solution (PSB), Chloroauric acid (HAuCl4), sodium citrate (Na3C6H5O7), potassium carbonate (K2CO3), Au NPs, Membrane Hi-Flow™ Plus HF180 and C083 Cellulose Absorbent Sample Pads acquired from Sigma-Aldrich were used without further treatment. A QuantiTect Reverse Transcription kit and RNeasy Power Microbiome extraction kit were obtained from Qiagen (Hilden, Germany). A 0.45 nm pore diameter nitrocellulose membrane was purchased from Millipore (Burlington, MA, USA). Bst 3.0 enzyme, Bst buffer, and MgSO4 were purchased from New England Biologicals (NEB, MA, USA). dNTPs mix 10 mM, nuclease-free water and SYBR Safe DNA gel stain were obtained from Thermo Fisher Scientific (Waltham, MA, USA). HCl, tris acetate buffer (TAE 10X) was purchased from Sigma-Aldrich (St Louis, MO, USA). All chemicals were of analytical grade. Aqueous solutions were prepared with Milli-Q water.
Extraction and concentration of SARS-CoV-2 RNA
Wastewater samples were taken from the effluent in the region of Juriquilla, Querétaro since June 2022 to February 2023. These samples were concentrated using the electronegative membrane method due to its high detection limit for SARS-CoV-2 genes. The pH of the sample was adjusted to 3.5 with 2 N HCl, and the samples were filtered through a negatively charged nitrocellulose membrane with pores of 0.45 μm in diameter (Millipore, Netherlands). The membranes were cut and used in an RNA extraction kit, Power kit RNeasy Power Microbiome kit (Qiagen, Germany).45,46
RT-LAMP assay
First, we performed reverse transcription for 5 µL of RNA using a QuantiTect Reverse Transcription Kit (Qiagen, Germany) according to the manufacturer's instructions. We then prepared the RT-LAMP reaction of 50 μL, each reaction containing 3 μL MgSO4 (NEB), 5 μL Bst buffer (NEB), 5 μL 2 mM dNTPs (Thermo Scientific), 2 μL 10X primer core mix, 2 μL 10X primer loop mix, 10 U Bst 2.0 DNA polymerase (NEB), 4 μL cDNA, and nuclease free water, which was added to complete the total volume of 50 μL. We designed the specific primers for the SARS-CoV-2 N gene (see Table 1), using the tool PrimerExplorer V5 (https://primerexplorer.jp/e/). The RT-LAMP reactions were verified using standard electrophoresis in 1% agarose gel with SYBR Safe DNA Gel Stain to verify the correctness of the amplification products. Lastly, the RT-LAMP amplicons were validated through sequencing on the data base of ncbi (https://www.ncbi.nlm.nih.gov/nuccore/om522662).
The main advantages of LAMP method compared with conventional RT-PCR are the follow: 1) The LAMP technique, like PCR, can amplify specific fragments of DNA which allows highly sensitive detection of pathogens. 2) The effectiveness of LAMP is based on the design of primers that must be very specific. 3) Unlike PCR, LAMP requires 4 to 6 primers. 5) The method uses a DNA polymerase (BST enzyme) with strand displacement activity and, with Retrotranscription (RT) activity. This displacement amplification property does not require PCR step to denature (90°C). 6) It is isothermal, that is, the reaction occurs at the same temperature 60°C continuously, which is an important advantage over traditional PCR. 7) High amplification efficiency: DNA is amplified 109 – 1010 times in 15 to 75 minutes. 8) One of the great advantages is related to greater tolerance to inhibitors and less infrastructure and time to perform the reactions and obtain the results at molecular level.
Dipstick for Lateral Flow assay (LFD)
To detect LAMP products by LFD, FIP was 5′-labeled with biotin and a fluorescein isothiocyanate (FITC)-labeled (LF) probe for use in subsequent LFD assay steps. The biotinylated RT-LAMP product hybridizes with a FITC-labeled probe specific for the target DNA. This was designed between primers B1c and B2 for molecular hybridization detecting FITC-biotinylated LAMP product. Upon application to the dipstick, the hybridization product complexes with gold-labeled anti-FITC specific antibodies. The immune complexes continue to diffuse through the analytical membrane. By overflowing the test band with biotin-specific molecules immobilized on the test band, the labeled immune complexes are captured and produce a red test band over time. Labeled immune complexes are not captured at the test line but overflow the control band and are immobilized by species-specific antibodies. An intense control band is generated over time. Primer sequences are shown in Table 1. According to the LFD test protocol (Milenia Genline HybriDtect, Germany), after completion of the biotin-labeled RT-LAMP reaction without heat inactivation, 8 μL of RT-LAMP reaction product (hybridized product) was added to 100 μL of detection buffer in a new tube. Finally, the LFD strip was immersed in the mixture. Detection results were determined by observing the control and test lines on the LFD strips.
Determination of RT-LAMP-LFD specificity
To determine the specificity of our system design and procedure, we performed RT-LAMP assays by annealing the seasonal H1N1 influenza genome and LAMP assays with DNA from A549 cells, then we confirmed the results on a 1% agarose gel, data not shown.
Evaluation of RT-LAMP-LFD sensitivity
LAMP-LFD sensitivity was evaluated against 10-fold serial dilutions of a positive control template (synthetic N gene of SARS-CoV-2). These dilutions ranged from 1 copy to 1000 copies, with nuclease-free water included as a negative control (NTC). To validate our results, each concentration was tested three times. We also performed a RT-LAMP colorimetric assay, reactions were performed in 25 μL reaction volumes.
Applicability of the RT-LAMP-LFD assay to field samples
The 12 water samples collected from Queretaro, Mexico were assayed using LAMP-LFD and the results were compared with those obtained using conventional colorimetric RT-LAMP assays. Ultrapure water was used instead of RNA template in the negative control. The results of the standard RT-LAMP reaction were analyzed by 1% agarose gels and sequencing. RT-LAMP reaction products were analyzed using LFD strips.
LAMP-Lateral Flow assays
To validate RT-LAMP assays that amplify the N gene of the SARS-CoV-2 virus, we first performed a series of dilutions using a synthetic N gene in colorimetric RT-LAMP assays as shown in Figure 1. The colorimetric RT-LAMP reaction produces a magenta to yellow color change when the gene of interest is amplified. The RT-LAMP reaction was performed at 63°C for 30 minutes using the primers listed in Table 1. As shown in Figure 1, our RT-LAMP assay achieves up to one copy of the synthetic N gene.
|
|
F3 |
CCAGAATGGAGAACGCAGTG |
0.2 |
|
B3 |
CCGTCACCACCACGAATI |
0.2 |
|
|
N |
FIP |
AGCGGTGAACCAAGACGCAG- |
1.6 |
|
GGCGCGATCAAAACAACG |
|||
|
Amplicon: |
BIP |
AATICCCTCGAGGACAAGGCG- |
1.6 |
|
AGCTCTTCGGTAGTAGCCAA |
|||
|
215 pb |
F2 |
GGCGCGATCAAAACAACG |
0.8 |
|
Flc |
AGCGGTGAACCAAGACGCAG |
0.8 |
|
|
B2 |
AGCTCTTCGGTAGTAGCCAA |
0.8 |
|
|
|
Blc |
AATICCCTCGAGGACAAGGCG |
0.8 |
Table 1 RT-LAMP primers for the detection of SARS-CoV-2 N gene from wastewater samples
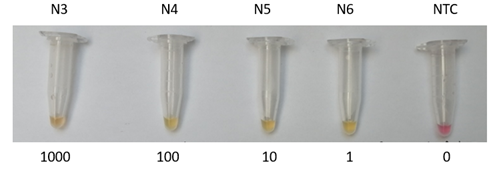
Figure 1 End-point measurements to detect RT-LAMP products of N genes. (a) Output characteristic curves of synthetic N gene, N3=1000 copies of N gene, N4=100 copies, N5=10 copies, N6=1 copy and negative template control (NTC) samples.
RT-LAMP-LFD sensitivity
After setting up the RT-LAMP conditions with the synthetic N gene for the colorimetric reaction, we decided to determine the sensitivity of our RT-LAMP-LFD system by performing a series of dilutions using the synthetic N gene as a template. As shown in Figure 2, our RT-LAMP-LFD system has a detection limit of one copy of the N gene, similar to the results obtained with the colorimetric RT-LAMP reaction shown in Figure 1.
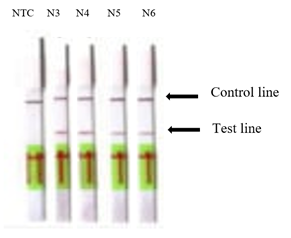
Figure 2 Sensitivity of RT-LAMP-LFD of SARS-CoV-2 using a synthetic N gene and no template control (NTC). Serial dilution of synthetic N gene; N3=1000 copies, N4=100 copies, N5=10 copies, N6=1 copy, respectively.
Water sample analysis with RT-LAMP-LFD assays
Twelve RNA samples from wastewater effluents from the area of Querétaro, Mexico, a negative control for the N gene and a positive control which is the synthetic N gene were analyzed by the RT-LAMP-LFD assay. All twelve field water samples were positive for the SARS-CoV-2 N-gene detected by the RT-LAMP-LFD assay (Figure 3). To ensure experimental accuracy, the positive samples were further analyzed by colorimetric RT-LAMP and sequencing of the RT-LAMP products. As expected, the results obtained by RT-LAMP-LFD are reliable, and this confirms us that this system is functional for the detection of SARS-CoV-2 virus N from wastewater samples.
The novel RT-LAMP-LFD method developed in this work can be used for the detection of the SARS-CoV-2 virus genome, in a time of 30 minutes (excluding the extraction and concentration step of the genetic material), at a constant temperature of 63°C. This device does not require expensive equipment or highly qualified personnel for its use, which makes it a methodology that can be used on site to assist in the work of environmental epidemiological monitoring.
This work was supported by the Project Support Program for Research and Technological Innovation (PAPIIT) of UNAM (Project TA100621).
The authors declare that they have no conflict of interest.

©2024 Castillo-Villanueva, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.