Journal of
eISSN: 2373-437X


Research Article Volume 11 Issue 1
1Bacterial resistance laboratory, Biomedicine department, IIBCAUDO, Universidad de Oriente, Venezuela
2Bioanalysis department, Universidad de Oriente, Venezuela
Correspondence: Abadía-Patiño L, Bacterial resistance laboratory, Biomedicine department, IIBCAUDO, Universidad de Oriente, Av. Universidad, Cerro del Medio, Cumaná, Estado Sucre, Venezuela, Tel +584148040684
Received: January 20, 2023 | Published: January 27, 2023
Citation: Abadía-Patiño L, Bastardo MF. Contamination with Enterococcus spp., strains of lettuces of different species acquired in Cumaná. J Microbiol Exp. 2023;11(1):13-18 DOI: 10.15406/jmen.2023.11.00380
Lettuce is one of the most common crops in the world, with high index of vitamins, minerals and fiber. Raw food consumption is closely related to foodborne illness, of which Escherichia coli O157:H7, seems to be the prime perpetrator leading to undesirable consequences. However, E. coli is not the only microorganism that can be transmitted, as reports validating the involvement of Aeromonas, Yersinia, Listeria, Staphylococcus, Campylobacter, and Salmonella have surfaced. The objective of this work was to detect the contamination of lettuces of different species acquired in several places of Cumaná, with Enterococcus strains. Of 52 lettuces acquired in food retail centers, 38% were contaminated with Enterococcus spp., strains. The best-selling lettuce was Lactuca sativa L. (25/52), but the most contaminated was Lactuca sativa var. capitata (89%). The highest average for Enterococcus was obtained in lettuces from Municipal Market. The dominant species of Enterococcus were E. faecalis (40%), and E. casseliflavus (30%); and to a lesser extent E. faecium (15%), E. gallinarum (10%), and E. avium (5%). The susceptibility profile showed that there are strains resistant to glycopeptides, fluoroquinolones, ansamycins, macrolides, phenols and tetracyclines. The clonal dissemination of two strains of E. faecalis and one strain of E. faecium by antibiotyping was demonstrated in lettuces from the municipal market. These results demonstrate that the food chain is a route of dissemination of multidrug resistant Enterococcus to the human intestinal microbiota, turning the gastrointestinal tract into a reservoir of intractable bacteria with the available antibiotics.
Keywords: fecal contamination, foodborne illness, bacterial resistance, free antibiotics wastewater, irrigation water, agriculture, potential risk
Several studies have shown that vegetables grown in developing countries may be contaminated with pathogenic microorganisms, and the scope of contamination is high when wastewater is used for irrigation. Other sources of potential risk to health are organic fertilizers, methods of transporting products, handling in markets and at points of consumption.
Hygienic measures are an important for food safety, especially for vegetables eaten raw that grow close to the soil surface contaminated with human and animal fecal matter which include poultry manure. The survival of these bacteria in the soil after manure application can be up to 100 days. The transfer of pathogens to lettuce can occur through splashing effects caused by raindrops, sprinkler irrigation or by means of transport of soil particles by mechanical weeding.1
The consumption of vegetables contaminated with pathogenic microorganisms, especially in urban areas where wastewater is used to irrigate vegetable crops, has been the prime reason for the outbreaks of public health manifestations. To reduce the risk associated with the consumption of contaminated vegetables, people must know vegetable decontamination methods.2 Increase in fast food consumption in street stalls, has been one of the main factor related to health problems associated with the proliferation of microorganisms due to unhygienic practices causing gastrointestinal problems,3 and of these foods, the most contaminated are vegetables (including lettuce). In Argentina, a study was carried out in which it was demonstrated that resistant Enterococcus strains that were believed to be confined to the hospital setting were found in the community and that lettuce could be the vehicle for transmitting these pathogens to healthy individuals.4
The purpose of carrying out this work was to detect the contamination of lettuce of different species acquired in different places of Cumaná, with strains of Enterococcus, which will contribute to define its possible role as a reservoir of resistance genes to antibiotics for human clinical use.
Samples
The samples were 52 lettuces of different species which were previously identified as Lactuca sativa L. (Batavia), Lactuca sativa var. longiofolia (Roman), Lactuca sativa var. capitata (Icerberg), Cichorium intybus var. foliosum (Red chicory) and Cichorium endivia var. crispum (Endive), were acquired in various commercial establishments in Cumaná, Sucre state, in Venezuela. Sampling was carried out following the table of random numbers.
Isolation and recovery of Enterococcus spp., from lettuce of different species
10 g of lettuce leaves were placed in tubes with Brain Heart Infusion (BHI) broth; then they were incubated in a shaking water bath at 35ºC for 24 hours. An inoculum was taken from each tube and plated on Enterococcus Confirmatory agar (Biomark); the plates were incubated at 35ºC for 24 hours.5
Colonies with typical morphology for Enterococcus (yellow or cream) were sought on the selective agar plates with growth visible. All colonies were tested for catalase to verify that they were catalase negative. The selected colonies on the plates were transferred with a wooden toothpick to hemolysis tubes with 1 mL of BHI broth with 6.5 NaCl and placed in a water bath at 45ºC for 24 hours to confirm the presence of Enterococcus.6
Identification of Enterococcus species
Once the presence of Enterococcus strains was confirmed, the species were identified through the RAPID ID 32 STREP gallery (bioMérieux, Marcy-l'Étoile, France), which consists of 32 tests for streptococci and bacteria similar to this family. From a pure culture, a suspension of the strains was prepared at a turbidity equivalent to 4 McFarland. The gallery domes were inoculated with 55 µL of the suspension. The galleries were incubated at 35°C for 4 hours and read according to the manufacturer's instructions.
Detection of antibiotypes of strains of Enterococcus spp., from lettuce through antimicrobial susceptibility tests
The antimicrobial susceptibility profile was performed using the disk diffusion method. An inoculum corresponding to the 0.5 McFarland pattern of each strain was prepared, Müeller-Hinton (MH) agar plates were sown, according to the instructions in the M100-S26 manual. The antibiotics used were: vancomycin (30 µg), teicoplanin (30 µg), ampicillin (10 µg), chloramphenicol (30 µg), erythromycin (15 µg), tetracycline (30 µg), ciprofloxacin (5 µg), linezolid (30 µg), norfloxacin (10 µg), nitrofurantoin (300 µg), rifampin (5 µg). They were incubated at 35ºC for 24 hours, then the inhibition zones were read and correlated with the 2D interpretive tables published in the M100-S26 manual.7
The antibiotyping was carried out by comparing all the strains of the same species, from the point of view of the profile of the antimicrobial susceptibility tests, in order to distinguish whether the Enterococcus strains found in the lettuce were from the same strain or was it a different one.
Minimum inhibitory concentration
The minimum inhibitory concentration (MIC) to ampicillin, vancomycin and ciprofloxacin was determined by the MH agar dilution method, according to the standards of the M100-S26 manual.7 The strains used for quality control were: S. aureus ATCC 25923 (the negative control) and E. faecalis 77904 VanB (positive control). Agar plates containing different concentrations of antibiotic (0.5 – 512 µg/mL) were inoculated and incubated for 24 hours at 35ºC. The CMI is the first concentration where there is no visible growth. The presence of 1 to 3 colonies was not taken into account.
Statistical analysis
Once the necessary assumptions for the data obtained were verified, an analysis of variance (95%) was applied in order to obtain the possible existence of a difference in the frequency of the bacterial group between the types of lettuce, commercial warehouses and the times of sampling. The results obtained were presented in the form of tables and figures.8
Fifty-two lettuces purchased in various commercial establishments in the city of Cumaná were studied. In the Municipal Market, five Lactuca sativa L, three Lactuca sativa var. longiofolia and three Lactuca sativa var. capitata; from the Mobile Stand: 16 Lactuca sativa L., from the Supermarket on the outskirts of the city: two Lactuca sativa var. capitata and two Cichorium intybus var. foliosum, from the Supermarket in the city center: four Lactuca sativa L. and two Lactuca sativa var. capitata, from the most popular supermarket: two Lactuca sativa var. capitata, from the exclusive Supermarket: three Lactuca sativa var. longiofolia and two Cichorium endivia var. crispum, from the new supermarket: three Cichorium intybus var. foliosum and five Cichorium endivia var. crispum.
Of the total lettuce studied, 20 were positive for Enterococcus (38%), of which the same number of strains of different species was obtained. According to the results obtained, it can be seen in table 1 that the highest frequency of isolation of Enterococcus spp., occurred in Lactuca sativa var. capitata (89%; 8/9), followed by Cichorium intybus var. foliosum (60%; 3/5). The lettuce acquired in the exclusive establishments and the street stall were not contaminated with Enterococcus.
Lettuce |
Nº of samples |
Nº strains isolated |
Frequency (%) |
Lactuca sativa L. |
25 |
4 |
16 |
Lactuca sativa L. var. longifolia |
6 |
3 |
50 |
Lactuca sativa var. capitata |
9 |
8 |
89 |
Cichorium intybus var. foliosum |
5 |
3 |
60 |
Cichorium endivia var. crispum |
7 |
2 |
28 |
Table 1 Frequency of Enterococcus spp. isolations, according to the type of lettuce studied
Nº, number; %, percentage
When applying the simple ANOVA test to compare the amount of bacteria identified according to the place of purchase (Figure 1), significant differences were detected (p <0.05), observing the highest average of Enterococcus in lettuce from the Municipal Market and the minimum average in the street stall and the exclusive supermarket. The a posteriori test shows the formation of two independent groups, presenting a higher average for the group represented by lettuce from the municipal market.

Figure 1 Analysis of Variance (ANOVA) of the lettuce retail centers studied and the frequency of Enterococcus isolates.
MM, municipal market; PA, street stall; SC, supermarket in the city center; SE, exclusive supermarket; SP, supermarket on the outskirts of the city; SMP, most popular supermarket; SN, new supermarket
In the results obtained in general, the Enterococcus species mostly isolated in the lettuce samples were E. faecalis (40%) and E. casseliflavus (30%); while E. faecium (15%), E. gallinarum (10%) and E. avium (5%) were the least frequent.
The Enterococcus species recovered in the Lactuca sativa L. samples were E. faecalis (75%) and E. faecium (25%). Regarding Lactuca sativa L. var longifolia, only the species E. faecalis (100%) was isolated. In Lactuca sativa var. capitata, the most frequently isolated species was E. casseliflavus (75%), followed by E. gallinarum and E. faecium (12.5% respectively).
When applying the simple ANOVA test to compare the number of Enterococcus identified according to the lettuce class (Figure 2), no significant differences were found (p>0.05). This is due to the fact that most types of lettuce present similar averages in terms of the frequency of isolated bacteria.
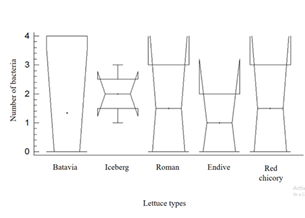
Figure 2 Analysis of Variance (ANOVA) of the types of lettuce studied and the frequency of Enterococcus isolates.
Batavia, Lactuca sativa L; Iceberg, Lactuca sativa var. capitata; Roman, Lactuca sativa var. longifolia; Endive, Cichorium intybus var. foliosum; Red chicory, Cichorium endivia var. crispum
In Cichorium intybus var. foliosum, three species (E. faecalis, E. gallinarum and E. avium) were isolated with the same frequency (33.3%). In Cichorium endivia var. crispum two species were isolated, E. faecalis and E. faecium in equal percentage (50%).
Table 2 shows the antimicrobial susceptibility profile of the Enterococcus faecalis strains. The strains show resistance to fluoroquinolones (ciprofloxacin and norfloxacin), macrolides (erythromycin), and ansamycins (rifampicin).
N |
AMP |
VAN |
TEC |
CIP |
NOR |
LZD |
ERY |
TET |
C |
RIF |
F/M |
1 |
S |
S |
S |
R |
R |
S |
I |
S |
R |
R |
S |
3 |
S |
S |
S |
I |
S |
S |
I |
S |
S |
S |
S |
1 |
S |
S |
S |
I |
I |
S |
I |
S |
S |
R |
S |
2 |
S |
S |
S |
S |
S |
S |
I |
S |
S |
R |
S |
1 |
S |
S |
S |
S |
S |
S |
R |
S |
S |
R |
S |
Table 2 Antibiotypes of Enterococcus faecalis strains isolated from different lettuces in Cumaná
N, number of strains; AMP, ampicillin; VAN, vancomycin; TEC, teicoplanin; CIP, ciprofloxacin; NOR, norfloxacin; LZD, linezolid; ERY, erythromycin; TET, tetracycline; C, chloramphenicol; RIF, rifampicin; F/M, nitrofurantoin
Table 3 shows the antimicrobial susceptibility of E. casseliflavus present in the samples of Lactuca sativa var. capitata. According to these results, these strains are 100% sensitive to the antibiotics teicoplanin and nitrofurantoin, but 100% resistant to rifampicin with intermediate susceptibility to erythromycin. Two strains resistant to linezolid.
N |
AMP |
VAN |
TEC |
CIP |
NOR |
LZD |
ERY |
TET |
C |
RIF |
F/M |
1 |
S |
S |
S |
S |
S |
S |
I |
I |
I |
R |
S |
1 |
S |
S |
S |
I |
I |
S |
I |
S |
S |
R |
S |
1 |
R |
I |
S |
R |
R |
R |
I |
S |
S |
R |
S |
1 |
S |
S |
S |
I |
I |
S |
I |
S |
S |
R |
S |
1 |
S |
I |
S |
I |
I |
S |
I |
S |
S |
R |
S |
1 |
S |
S |
S |
I |
S |
R |
I |
S |
I |
R |
S |
Table 3 Antibiotypes of Enterococcus casseliflavus strains isolated from different lettuces in Cumaná
N, number of strains; AMP, ampicillin; VAN, vancomycin; TEC, teicoplanin; CIP, ciprofloxacin; NOR, norfloxacin; LZD, linezolid; ERY, erythromycin; TET, tetracycline; C, chloramphenicol; RIF, rifampicin; F/M, nitrofurantoin
Table 4 shows the antimicrobial susceptibility of E. gallinarum isolated from Lactuca sativa var. capitata, Cichorium intybus var. foliosum and Cichorium endivia var. crispum in which 100% resistance to rifampicin can be observed, as well as intermediate susceptibility to fluoroquinolones, linezolid, erythromycin and tetracycline. Regarding the clonality of the strains, it was observed that in the E. faecalis species there are three clonal strains, two from Lactuca sativa L. (9E5 and 9E8) and one from Lactuca sativa L. var. longifolia (9E10), from three different lettuces, bought in different stalls on the same day in the Municipal Market. Two more strains of E. faecalis (9E6 and 9E9), one of Lactuca sativa L. and one of Lactuca sativa L. var. longifolia, also from the Municipal Market of the same day, but with another antibiotype. Regarding the species E. faecium, two clonal strains (9G8 and 11H8) of Lactuca sativa L. var. capitata were isolated on different days. and Lactuca sativa L. It is worth noting that the clonal strains were only in the lettuce purchased in the Municipal Market, which suggests that the source is the same, that is, it is a single supplier of lettuce for the vendors of the Cumaná Market, Sucre state. Table 5 shows that the MIC of ampicillin against Enterococcus oscillates between 1 and 8 μg/mL, with which they are considered sensitive; the only resistant strain is E. faecium. Regarding the vancomycin MIC, the bulk of the population is in the sensitivity range (between 0.5 and 4 μg/mL).
|
|||||||||||
N |
AMP |
VAN |
TEC |
CIP |
NOR |
LZD |
ERY |
TET |
C |
RIF |
F/M |
1 |
S |
R |
S |
R |
I |
R |
I |
I |
S |
R |
S |
1 |
S |
S |
S |
I |
S |
I |
I |
S |
S |
R |
S |
1 |
S |
S |
S |
I |
I |
S |
I |
S |
S |
R |
S |
Table 4 Antibiotypes of Enterococcus gallinarum strains isolated from different lettuces in Cumaná
N, number of strains; AMP, ampicillin; VAN, vancomycin; TEC, teicoplanin; CIP, ciprofloxacin; NOR, norfloxacin; LZD, linezolid; ERY, erythromycin; TET, tetracycline; C, chloramphenicol; RIF, rifampicin; F/M, nitrofurantoin
CMI |
0,5 |
1 |
2 |
4 |
8 |
16 |
32 |
64 |
128 |
VAN |
5 |
1 |
6 |
5 |
2 |
0 |
0 |
1 |
0 |
CIP |
10 |
0 |
2 |
4 |
3 |
0 |
1 |
0 |
0 |
AMP |
7 |
0 |
3 |
3 |
6 |
0 |
0 |
0 |
1 |
Table 5 Minimum inhibitory concentration (µg/ml) of Enterococcus spp. strains, isolated from lettuce samples studied
MIC, minimum inhibitory concentration; NPV, vancomycin; CIP, ciprofloxacin; AMP, ampicillin
In industrialized countries, the daily consumption of fresh, ready-to-eat vegetables has increased in recent years, health concern. Today, ready to use bagged lettuce or mixed vegetables are available in supermarkets, to eat salads without having to prepare them at home, just by uncovering a bag and adding a little dressing. With this practice, a greater morbidity of foodborne diseases has arisen, especially these raw.9
Plants, as well as animals and humans, have their own microbiota. In a study carried out on Lactuca sativa var. longifolia found that the most predominant phyla of microbiota bacteria are: Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria. Among the main genera that comprise the nucleus of the composition of the phyllospheric microbiota of lettuce are: Arthrobacter, Bacillus, Massilia, Pantoea and Pseudomonas. The composition of the bacterial community on the surface of lettuce leaves varies according to the time of year in which they are cultivated as a function of time, space and environment.10
These habitual plant colonizing microorganisms are not harmful to humans. The problem arises when other microorganisms contaminate plants with irrigation water, manure, cross contamination with animals, dirty equipment or human manipulation.11
Not finding contamination by Enterococcus in the lettuce purchased at the street stall and the exclusive supermarket, it could be because they come from places where they do not use wastewater for irrigation or organic matter for compost, but it could also be that the vendors wash well lettuce before putting it on display for sale.
The lettuces purchased in the municipal market and the new supermarket were the most contaminated, and may have acquired this contamination in the farmland, due to the aforementioned factors, but also due to incorrect handling techniques. In a study carried out in Corrientes, Argentina, contamination by Enterococcus strains (42%) was demonstrated in 79 samples of Lactuca sativa acquired in three different centers.5 In this investigation, it can be observed that Lactuca sativa L. is the most predominant; the one that was most contaminated by Enterococcus was Lactuca sativa var. capitata with 89% frequency (Table 1), acquired in seven food outlets in Cumaná, Sucre state.
The frequency of microorganisms in vegetables reflects the poor sanitary quality of the raw product at the time of consumption.12 In Argentina, 42% Enterococcus strains were isolated from lettuce samples and among the most frequent species were E. faecium and E. faecalis.5 In a study carried out in Côte d'Ivoire, both from lettuce (36 samples) and irrigation water from these crops (36 samples), 27 Enterococcus strains were isolated from lettuce and 29 from irrigation water. The main species was E. faecalis (75%), as well as strains of E. faecium, E. gallinarum, E. casseliflavus and E. durans.13 Regarding the present work, E. faecalis was always the predominant species in these foods.
Most Enterococcus infections in humans are caused by E. faecalis, however, E. faecium is responsible for serious infections by multidrug-resistant strains and is the species that causes most deaths. Infections caused by other Enterococcus species are not very frequent and do not present multi-resistance problems either.14 The results of this study reveal that most of the lettuces are colonized with E. faecalis as indicated by the international literature. Although E. faecium was not the predominant species, three strains resistant to all the antibiotics tested were isolated.
Regarding the susceptibility profile of the E. faecium strains isolated from fresh produce in the United States, they presented a high level of resistance to ciprofloxacin, tetracycline, and nitrofurantoin. In total, 91% of these strains were resistant to at least one antibiotic tested, as were 32% of the E. faecalis strains. These strains of E. faecalis, of the same origin, had a lower prevalence of resistance to human clinical antibiotics and those of agricultural relevance.15
These data reflect the serious problem of transmission of antibiotic-resistant pathogens through the food chain. In this work, all E. faecalis strains are 100% sensitive to nitrofurantoin. In the event that a person acquires this strain through the food chain and develops a urinary infection due to said strain (with a starting point due to fecal contamination), it is possible to treat them with nitrofurantoin. The fact that food is the vehicle for the transmission of Enterococcus to humans, that is, the reservoir for the horizontal transfer of resistance determinants between environmental and human strains, is a public health problem, since this species survives by passing the gastric barrier, multiplies and colonizes the intestinal tract, for long periods.15 Patients with predisposing factors can develop infections by colonizing strains of their intestine, with mechanisms of resistance to antibiotics for human clinical use.
In countries like Argentina, Chile, and even Venezuela, there are very few studies or antecedents of antibiotic susceptibility of Enterococcus that have been carried out on strains from food, with the exception of those that cause gastrointestinal infections; For this reason, it is very difficult to compare these results. Continuous surveillance is necessary to know if the transmission of these bacteria occurs via the food chain or is generated in the environment.16
Immunosuppressed patients are those who can develop hospital-acquired infections due to strains of E. gallinarum and E. casseliflavus, causing bacteremia, endocarditis, septicemia, or urinary tract infection.17 The danger of acquiring an infection from one of the strains isolated in this work is that some are resistant to antibiotics for human clinical use, as is the case with a strain of E. faecalis resistant to erythromycin, tetracycline, chloramphenicol, and rifampicin. More worrisome is that strains of E. casseliflavus are resistant to fluoroquinolones, linezolid, and ampicillin. This strain must be studied to determine if it produces penicillinase, since a difference greater than 5 mm was observed when the ampicillin-sulbactam disk was placed, compared to the ampicillin disk. They are usually colonizing strains and are uncommon in human infections.18
E. gallinarum and E. casseliflavus are not frequently isolated in clinical samples, but it must be recognized that they can cause severe invasive disease. Clinical experience with such strains has been limited. A review of the literature reveals that E. gallinarum or E. casseliflavus/flavescens can be isolated from a variety of patients who are chronically ill or immunosuppressed. Mainly, they are isolated from cases of bacteremia in patients with underlying conditions, such as renal failure, diabetes mellitus, hematological malignancy, organ or bone marrow transplant recipients, among other conditions.18
Three strains of E. faecium were isolated in this work; each of different lettuces (Lactuca sativa L., Lactuca sativa var. capitata, Chichorium endivia var. crispum) resulting in high-level resistance to all antibiotics tested, while the only E. avium strain was totally resistant to linezolid and rifampicin, with intermediate sensitivity to ciprofloxacin, erythromycin and chloramphenicol. The latter is reflected in a study in which strains of E. faecium isolated from vegetables, soil, farm animals and manure were analyzed. Thirty-five strains out of 37 were resistant to aminoglycosides at a high level, confirmed by the CMI and by the detection of the resistance genes [aac(6')-Ie-aph(2”), aph(2')- Ic, aph(3')-IIIa and ant(4')-Ia].1
In a study with food of animal origin (shrimp), strains of E. faecium (49%), E. faecalis (29%), E. gallinarum (10%), E. hirae (5%), E. casseliflavus (3%), and E. durans (3%).19 The strains were resistant to vancomycin (37%), tetracycline (46%) and erythromycin (49%), similar to what occurred in this study. In this study it was not possible to demonstrate the susceptibility profile of the isolated strains to aminoglycosides, due to the non-availability of high-load disks to these antibiotics in the country.
It is interesting to highlight that, in the literature, they refer to the importance that Enterococcus has acquired in recent times with respect to the high level of resistance to aminoglycosides, since they are intrinsically resistant at a low level. For example, in one study16 they detected strains of E. faecalis (68%) and E. faecium (88%) resistant to gentamicin and both species 100% resistant to streptomycin, an important finding, since these strains were recovered from sewage, hence the importance of studying environmental bacteria and their resistance profiles.
Once these strains are consumed through the food chain, they are able to survive stomach acidity and pass to the gut to colonize it, becoming a human reservoir of potentially pathogenic strains, resistant to antibiotics for human clinical use, to treat serious infections.
This colonization can last from several months to a year and when the conditions are right, self-infect with gut strains,20 such as long hospital stays, use of third-generation cephalosporins, hospitalization in intensive care, transplant patients, hematological disorders.21 Enterococcus translocation occurs when an overgrowth of these happens in the gut lumen, which is very frequent in patients undergoing antibiotic treatments that are not effective on this genus,22 and can perfectly cause serious infections. The results obtained in this investigation are of great relevance for public health, since Enterococcus strains with high percentages of resistance to antibiotics for human clinical use were found in bacteria from lettuce, a food widely consumed by the population.23
The food chain is a route of dissemination of multiresistant enterococci to the human gut microbiota, turning the gastrointestinal tract into a reservoir of bacteria that is intractable with available antibiotics.
None.
The authors of the work declare that they have no conflict of interest with the distribution centers of lettuces.

©2023 Abadía-Patiño, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.