Journal of
eISSN: 2373-437X


Research Article Volume 1 Issue 1
1College of Veterinary Medicine, Mississippi State University, USA
2Biological Sciences, Mississippi State University, USA
3Amity Institute of Biotechnology, Amity University, India
Correspondence: Mark L Lawrence, College of Veterinary Medicine, Mississippi State University, Mississippi State, MS-39762, USA
Received: April 14, 2014 | Published: April 24, 2014
Citation: Paul D, Scott J, Green M, et al. Composition of bacterial communities isolated from core samples taken from petroleum deposits. J Microbiol Exp. 2014;1(1):1-7. DOI: 10.15406/jmen.2014.01.00001
Microbial communities in the subterranean environment are unique in that they are completely isolated from biological communities that rely on photosynthesis, earth's atmosphere or oceans. Interestingly, bacteria found in petroleum deposits are often in a dormant form because petroleum deposits are very limited in nitrogenous and phosphorous-containing nutrients. These are unique populations as compared to other subsurface microbial communities and might consist of novel species, new metabolic capabilities, and undiscovered adaptive microbial mechanisms. The goal of the current study was to isolate and identify cultivable bacteria (mainly oligotrophs) and develop a method to evaluate bacterial community structure in petroleum deposits using 16S rDNA based technique.
Keywords: petroleum deposits, bacterial communities, ARDRA and sequencing
Bacteria can exist under high pressure and temperature conditions, in low oxygen or low nutrient conditions, and sometimes are able to remain dormant for several years. Oilfields usually represent extreme environments, poor in nitrates and oxygen, but harbor a diverse community of sulfate-reducing, methanogenic and fermentative bacteria, and hydrocarbon or oil-utilizing bacteria. Some of the important edaphoclimatic factors that influence microbial community composition and diversity include pH,1 particle size,2 organic carbon,3 nitrogen/phosphate,4 water,5 and oxygen.6 The physiology and metabolic potential of microbial communities will vary greatly with location along a soil profile depending on the available water, plant derived resources (carbon, nitrogen, and other nutrients), mineralizable carbon and nitrogen, and oxygen. In mines and subsurface soils, these factors cause selective pressures that tend to decide the nature and distribution of bacteria, especially oxygen/nutrient limitations and temperature/pressure.7 The most widely represented and best-known types are sulfate-reducing, methanogenic, and fermentative bacteria.8–11
Some of the bacteria in mines and petroleum deposits survive the extreme environment by maintaining a dormant state.8 These bacteria tend to behave like oligotrophic bacteria10 or unculturable bacteria when grown in lab conditions because of long-term starvation conditions. Oligotrophic bacteria are slow-growing and ubiquitously occur in water or soil lacking organic substances.
To explore bacterial diversity in various environments/niches, 16S rRNA amplification and sequencing methods are commonly and efficiently used.12–15 Amplified ribosomal DNA restriction analysis (ARDRA)12 is one of the simplest and most cost-effective techniques for culture-independent description of bacterial communities.16 For several environmental samples such as soil or seawater, the proportion of cells that can be cultured is estimated to be 0.1% or at most 10% of the total population,17 and few data are available concerning how closely they reflect the actual composition of these communities. ARDRA allows ‘genetic fingerprinting' of the bacterial community dwelling in any particular environment and thereby provides a global picture of the genetic structure of the bacterial community.
Identification and quantification of petroleum reservoir microorganisms, including nitrate reducing bacteria (NRB) and sulfate reducing bacteria (SRB), has been assessed by cultivation-dependent methods,18 and cultivation-independent methods have only recently been introduced into the field of reservoir microbiology.19,20 Considering the small number of these studies, information currently available on the microbial communities present in oil reservoirs is sparse and, most notably, the microbial diversity of drilling sample cores has never been studied before, probably due to lack of accessibility and difficulties in extracting bacteria/DNA from them. Most of the research so far dealt with production water from oil fields, oil dumps, or from the vicinity of oil.19,21–23
In the current study, we evaluated the microbial diversity directly from cores taken from an active oil well. This research is a good example of collaborative efforts between academia and industry. As a result of this study, we isolated and identified some of the hydrocarbon utilizing oligotrophic bacteria in petroleum reservoirs. This study required developing methods for bacteria and DNA extraction from petroleum core samples, and to our knowledge it is the first report to use culture independent study of bacterial communities from petroleum communities using ARDRA and sequencing. We intend to compare our results with similarly deciphered communities from other core samples (varying depths and regions) in the future.
Sample collection
Cores were obtained from approximately 700m below surface in an oil-bearing formation in Brookhaven, Alabama. Each core was stored in a BBL Gaspak container to keep it in anaerobic conditions at low temperatures (0-4°C). Each core was cut into 4 inch sections using sterile steel under an atmosphere of nitrogen. The center of the core was placed in a sterile stainless steel crusher and then sieved.
DNA extraction from the core samples
DNA was isolated directly from crushed core for metagenomic analysis. The PowerMax Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA) was adapted for use with a Barocycler (Pressure Biosciences, South Easton, MA). One gram of the crushed core material was vortexed with Power Bead buffer then mixed with lysis buffer and vortexed. The PCT (Pressure Cycling Technology) tube was loaded into the PCT machine and run for 80 cycles (35kpsi for 20sec, atmospheric pressure 5sec). This helped in lysis of the bacteria using high pressure. DNeasy spin column (QIAGEN) was used to purify the DNA from cell debris. The DNA was quantified and diluted with water for PCR.
PCR and cloning
Universal primers 27F (E. coli numbering 8~27; 5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (E. coli numbering 1492~1510; 5'-GGTTACCTTGTTACGACTT-3') (Cheneby et al., 2000) were used for amplification of bacterial 16S rRNA genes. PCR amplification of nearly ~500bp of 16S rRNA gene was performed in 25μl reaction mixtures. Reaction tubes contained 25 ng DNA, 1UTaq DNA Polymerase (New England Biolabs, Beverly, MA), 1Xbuffer (10mM Tris-HCl [pH 9.0], 1.5mM MgCl2, 500mM KCl, 10mM deoxynucleoside triphosphate, and 20pmol of each primer ml-1. Initial DNA denaturation and enzyme activation steps were performed at 95°C for 30s, annealing at 50°C for 1min, and extension at 72°C for 2min, with a final extension for 10min at 72°C. The presence and yield of PCR product was monitored by 1% agarose gel electrophoresis at 200V for 1h in 1X Tris-acetate-EDTA buffer and stained with GELSTAR.
16S rRNA gene clone libraries were constructed for elucidating the bacterial population in the cores. For this purpose, 16S rRNA gene amplicons from five independent PCRs were pooled, ligated into the TOPO 2.1 plasmid vector (Invitrogen, Life technologies, CA) and transformed into Escherichia coli DH5 5α-T1R chemically competent cells according to the manufacturer's instructions. The transformed cells were plated on Luria–Bertani agar plates containing 100mg ml-1 ampicillin, 40mg ml-1 isopropyl-b-D-thiogalactopyranoside, and 40mg ml-1 5-bromo-4-chloro-3-indolyl-b-D-galactoside. White clones were cultivated to analyze their plasmid content, and about 200 bacterial clones containing inserts of the correct size (1.5kb) were stored in 10% glycerol at -80°C.
ARDRA and sequencing
Two restriction enzymes (RsaI and Sau3A) were selected for digestion of the clones based on in-silica digestion profile of a few clones. Digestions were performed for 4h at 37°C in 25µl reaction volumes containing 10µl of the PCR product solution, 2.5µl of the incubation buffer (1X) and 0.3µl of the restriction enzyme (New England Biolabs). The reaction products were run on a 2% agarose gel, and the restriction pattern of the clones was compared.
Various phylotypes were identified on the basis of similarity in the restriction digestion pattern of 192 bacterial clones. Rarefaction analysis24 was used to confirm that the number of clone types had been exhausted after screening all the individual bacterial clones. We are aware that any given RFLP pattern may represent sequences from multiple phylogenetic groups and may therefore not represent a true phylotype in the traditional sense. Therefore, we use the term phylotype to indicate groups for richness calculations. Phylotype richness (S) was calculated as the total number of distinct RFLP patterns in a core. The Shannon–Weiner diversity index25 was calculated as follows: H=S(ri)(log2r_i), where r represents the proportion of a distinct RFLP pattern relative to the sum of all distinct patterns. Evenness was calculated from the Shannon–Weiner diversity index: E =H/Hmax where Hmax= log2(S). Partial 16S rRNA gene sequences (at least 400 bp) of one representative member of each phylotype were obtained using M13F primer with the ABI PRISM Big Dye Terminator Cycle Sequencing kit and an ABI 310 automated DNA sequencer (Applied Biosystems, Foster City, CA). These sequences were used to construct phylogenetic trees to understand phylogenetic relations between the phylotypes.
Data analysis
Partial 16S rRNA gene sequences were initially analyzed using BLAST search (www.ncbi.nlm.nih.gov/blast/blast.cgi) and RDPII analysis software (www.ce.msu.edu/RDP/html/analyses.html). The sequences were submitted to the NCBI (National Centre for Biotechnology and Biotechnology Information), and GenBank accession numbers were obtained for all of them. The closest match and GenBank accession numbers of each sequence are listed in Table 1 & 2. Clustal X26 was used to align these sequences. Sequence dissimilarities were converted to evolutionary distances according to the method of Jukes and Cantor (1969). Dendrograms were constructed with a neighbor-joining algorithm using NJ PLOT.27
Isolate |
Gen bank ID |
Best hit BLAST ID |
Identity % |
Match |
BS2 |
JF963965 |
JF322796 |
98 |
Bacillus cereus |
BS65 |
JF963966 |
HM030754 |
99 |
P. strutzeri |
BS67 |
JF963967 |
GU565237 |
99 |
Pseudomonas sp. |
BS67A |
JF963968 |
GQ150491 |
94 |
Paenibacillus sp. nC2 |
BS71 |
JF963969 |
FJ817481 |
92 |
Bacillus sp |
BS78 |
JF963970 |
JF714216 |
98 |
Bacillus sp |
BS29A |
JF963971 |
FR851254 |
99 |
Bacillus sp. PR1.7 |
BS81A |
JF963972 |
AY308758 |
99 |
Paenibacillus faviospores |
BS66A |
JF963973 |
EU368479 |
99 |
Paenibacillus |
Table 1 Partial 16S rDNA sequences and GenBank numbers of some of the isolates from the petroleum deposit core material
Rep-clone |
Gen bank no. |
No. of Clones |
Identification |
Closest hit BLAST ID |
Identity % |
CVMbac7 |
JF922889 |
10 |
Caulobacter sp |
FJ870519 |
99 |
CVMbac8 |
JF922913 |
1 |
B proteobacterium |
HQ823674 |
99 |
CVMbac10 |
JF922890 |
2 |
Propionibacteria |
HQ585178 |
99 |
CVMbac13 |
JF922891 |
56 |
Sphingomonas sp. |
HM057723 |
99 |
CVMbac15 |
JF922892 |
6 |
Schlegelella sp |
FN667261 |
99 |
CVMbac23 |
JF922893 |
2 |
Flavobacterium |
FR691516 |
99 |
CVMbac35 |
JF922895 |
7 |
Streptococcus suis |
AY753315 |
98 |
CVMbac27 |
JF922894 |
14 |
alpha proteobacterium |
HM480175 |
98 |
CVMbac42 |
JF922896 |
7 |
alpha proteobacterium |
EU429493 |
99 |
CVMbac48 |
JF922898 |
1 |
Tepidimonas sp |
EU450347 |
48 |
CVMbac43 |
JF922897 |
1 |
Phenylobacterium |
JF202879 |
98 |
CVMbac49 |
JF922899 |
2 |
beta proteobacterium |
EU450314 |
98 |
CVMbac50 |
JF922900 |
9 |
Pseudomonas sp |
HQ660803 |
99 |
CVMbac77 |
JF922901 |
2 |
Gamma proteobacterium |
FR774582 |
100 |
CVMbac91 |
JF922902 |
6 |
Gamma proteobacterium |
DQ230964 |
99 |
CVMbac107 |
JF922903 |
5 |
alpha proteobacterium |
HM110470 |
99 |
CVMbac193 |
JF922912 |
1 |
alpha proteobacterium |
EU148878 |
92 |
CVMbac121 |
JF922904 |
1 |
alpha proteobacterium |
HM480264 |
99 |
CVMbac132 |
JF922905 |
3 |
alpha proteobacterium |
HM799072 |
97 |
CVMbac157 |
JF922908 |
6 |
beta proteobacterium |
EF648091 |
100 |
CVMbac166 |
JF922909 |
4 |
Actinobacterium |
JF346422 |
99 |
CVMbac184 |
JF922910 |
3 |
alpha proteobacterium |
AY293404 |
99 |
CVMbac135 |
JF922906 |
2 |
Schlegelella sp |
HM316462 |
91 |
CVMbac147 |
JF922907 |
1 |
Leptothrix sp |
HM186821 |
100 |
CVMbac189 |
JF922911 |
4 |
alpha proteobacterium |
EF658677 |
99 |
Table 2 GenBank accession numbers of representative clones for each phylotype along with GenBank IDs of the closest BLAST hit and % identity
Culturing some of the aerobic microorganisms (oligotrophs) for identification
Fifty grams of crushed core material was suspended in simulated production water,28 thoroughly mixed, and streaked on either Bacto-Tryptic Soy Agar (TSA) or Bacto-Plate Count Agar prepared with simulated production water containing the following per 8 liters of distilled water (NaCl 778.00g, Na2SO4 130.00g, MgCl2.6H2O 352.00g, CaCl2.2H2O 36.00g, KCl 11.00g, Na2HCO3 3.20g, KBr 1.60g, SnCl2.6H2O 0.67g, H3BO3 0.41g, Na2SiO3.9H2O 0.08g, NaF 0.05g, NH4NO3 0.03g, FePO4.4H2O 0.02g). Plates were also streaked on oil agar that was prepared with simulated production water supplemented with 0.1% KNO3, 0.37% K2HPO4. 3H2O, 1% filter-sterilized crude oil, and 2% Bacto-Agar. After the agar had been poured into a petri plate and allowed to harden, a thin overlay was added using oil agar prepared with oil-saturated water, but containing no added oil. After incubation, colonies picked into simulated production water, thoroughly mixed, and re-streaked on the same agar from which the colony came. After incubation, a single colony was picked onto an agar slant of the same agar from which the colony was obtained and incubated at 30°C until growth was evident. An appropriate control (without core material) was also serially diluted and plated. Pure individual colonies were picked up and stored at -80°C as glycerol stocks. Some petri plates were poured with 16% bacto agar containing 1/8Kth TSA for growing oligotrophs/bacteria that would only grow under low levels of nutrients.
Genomic DNA was isolated from cultured isolates using Wizard genomic DNA purification kit (Promega). DNA was run on 0.7% agarose gel, quantified, and used for PCR amplifications. For identification of bacterial colonies, the same PCR primers described for metagenomic analysis (universal primers 27F and 1492R) were used for amplification of bacterial 16S rRNA genes. The same PCR conditions were used as described above. The presence and yield of PCR product was monitored by 1% agarose gel electrophoresis.
Microbial diversity and ARDRA
DNA isolation from the cores was possible using the Barocycler (Pressure Biosciences) and the method described (Figure 1). This DNA was diluted 10times to obtain amplification using the universal primers as described. About 192 clones were screened for bacteria, out of which 156 showed the presence of 16S rRNA gene inserts. Based on the clarity of banding patterns with Sau3AI, HaeIII, and RsaI, Sau3AI digestion profiles were used for identifying the 25 distinct phylotypes in bacteria. Rarefaction analysis curve (Figure 2) indicated that the population diversity has been sufficiently covered. For this community, a Shannon's diversity index (H) was calculated of 3.61 and species evenness (E) of 77.7. The evenness values approximate the maximum possible values, as most of the sequence types were recovered only once. Representative members of each phylotype were partially sequenced, and a phylogenetic tree was constructed (Figure 3) to show the relationships among the various phylotypes. The affiliation of clone sequences to described taxa indicate the presence of mainly aerobic species belonging to Proteobacteria, although not always deducible with a high degree of confidence. Thirty-six clones did not have 16S gene and were chimeric or false positives.
Figure 1 Isolation of metagenomic DNA from crushed cores using a technique developed in our lab. The last well in the gel shows the DNA ladder 1 kb plus.

Figure 2 Rarefaction curve (no. of clones analyzed vs no. of phylotypes) indicating sufficient representation of bacterial diversity after screening about 200 clones. Clones were grouped as phylotypes on the basis of their RFLP pattern.
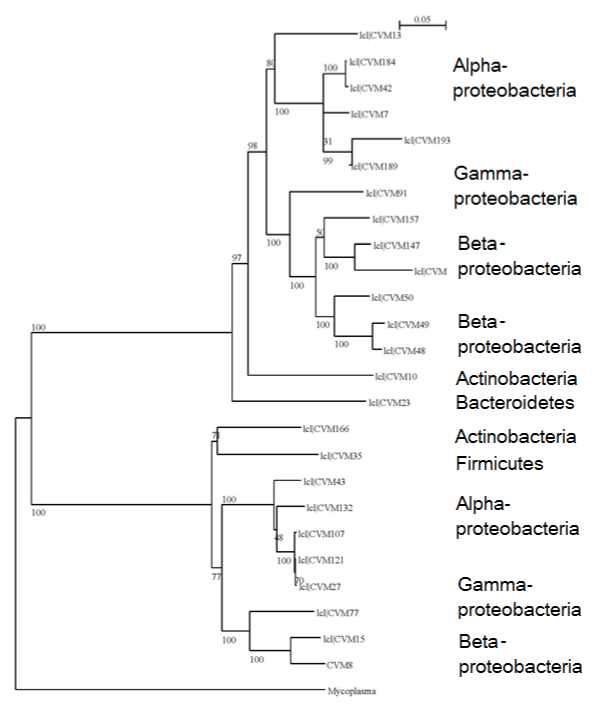
Figure 3Dendrogram showing the phylogenetic relationships between various bacterial phylotypes. Bootstrapping values indicate the strength of each node. Distance scale indicates evolutionary distance. Mycoplasma has been used as the out group to root the tree.
Various types of Alphaproteobacteria were abundant (67%) (Figure 4). These bacteria show high degree of identity to species existing in the Yellow Sea and microbial mats in hypersaline lakes. Presence and abundance of marine/aquatic bacteria is interesting because it might indicate their tolerance to high pressure conditions and extreme environments. Some bacteria from hypersaline lakes might be involved in exopolymer degradation (unpublished results). Some species identified are similar to endosymbionts or gut bacteria of insects. Also bacteria similar to those found in Panda feces indicate their capability to degrade plant material, possibly contributing toward petroleum formation. Caulobacter sp. was detected, which are prosthecate bacteria often specialized for oligotrophic environments.29
Gammaproteobacteria and some Actinobacteria were detected along with a few unculturable species similar to those found in activated sludge or areas of heavy metal (uranium) contamination, which can also be extreme environments with high selective pressures. Presence of hydrocarbon- degrading Actinobacteria has been reported in soil collected from the vicinity of active oil wells and oil dumps.22,30–32 Strains similar to composting bacteria and those similar to lactic acid producing bacteria indicate anaerobic reducers.
Apart from these, iron reducers (Bacteroidetes, Gammaproteobacteria) and nitrate reducers (Betaproteobacteria, Gammaproteobacteria) were found, which is similar to previous reports.14,33 Some sulfate reducing bacteria and fermentative bacteria were also detected in our study but in comparatively lower percentages, which might be related to the conditions in the reservoir or isolation technique.
Schlegelella sp. and Tepidimonas spp. were the only hyperthermophilic strains detected in the population. Some species of the genus Schlegelella have been found to degrade poly (3-hydroxybutyrate) in a few cases via a thermotolerant PHB polymerase enzyme.34,35 Tepidimonas spp. is a slightly hyperthermophilic bacterium having optimal growth temperature of about 50–55°C; some strains of this genus are known to produce useful proteases.36,37 Another important beta proteobacterium identified in our study belonged to Leptothrix spp (obligate lithotroph), which is also called “iron bacteria” because of its importance in biological oxidation of iron and manganese.38,39 The obligate strains of Leptothrix are reported to grow on trace amounts of metal oxides38 and copiously produce extracellular sheaths encrusted with iron oxyhydroxides. The presence of this genus in petroleum deposits indicates the presence of oxides of Fe or Mn in trace amounts.
The phylogenetic tree Figure 3 shows two major groups/clades, both of which have Alpha-, Beta-, and Gammaproteobacteria distributed according to their phylogenetic proximity. This indicates that the phylotypes within any of the broad class (e.g. Alphaproteobacteria) are widely divergent and might be comprised of novel species of microorganisms.
Microbial diversity amongst the cultured strains
Using oil agar as media, we distinguished 9 different bacterial isolates and cloned their 16S rDNA in E. coli for sequencing. It is probable that these strains were mostly surviving in an oligotrophic form40 and could be cultured only in 1/8Kth diluted TSB agar instead of the normal concentration. Table 1 lists all the species that were cultured either aerobically or anaerobically. BLAST results showed that mainly the clones showed a high degree of 16S rDNA sequence based on identity to Bacillus sp. or Paenibacillus sp. and Pseudomonas sp. These bacteria commonly occur in soil rhizospheres. Interestingly, BS65 was strongly identical to P. strutzeri strains isolated from production water of oil reservoirs. Another isolate BS67 might be a Pseudomonas strain showing strong identity to uncultured bacteria found in reservoirs. Cho & Giovannoni41 revealed that sporadically detected Gamma proteobacteria gene clones from seawater are part of a phylogenetically diverse constellation of organisms mainly composed of oligotrophic and ultra microbial lineages that are culturable under specific cultivation conditions.41 Pseudomonas is a member of Gammaproteobacteria, and there is definite evidence of ultramicrobial Pseudomonas lineages occurring in core samples.42 Kim & Crowley43 studied the microbial diversity in ca. 28,000-yearold samples of natural asphalts from the Rancho La Brea Tar Pits in Los Angeles, CA and found that the predominant bacteria in both pits were Gammaproteobacteria including Xanthomonadaceae (5 clones) and Pseudomonadaceae (5 clones). Culturing on 10% TSA plates showed the presence of yielded five distinct isolates of Pseudomonas spp., eight isolates of Bacillus spp., and five isolates of Citrobacter spp.43 Therefore, these results are also in accordance with our findings with regard to the uncultivable and cultural microbial diversity, where cultivable strains (on 10% TSA) were represented by relatively few strains. However, we found Paenibacillus sp. instead of Citrobacter spp.
Three of the strains belong to the genus Paenibacillus; BS81A shows strong sequence identity to P. favisporus, which has xylanophytic characteristics. P. favisporus sp. nov. produces a wide variety of hydrolytic enzymes, such as xylanases, cellulases, amylases, gelatinase, urease and β-galactosidase. This species might have helped in the formation of oil from plant residues. Presence of oligotrophic Bacillus sp. is expected44 because they secrete a large amount of mucus and have a viscid capsule or thick pectic exine around themselves to survive extremities. Polyphasic identification and characterization of these isolates are currently underway. We are also trying to isolate several other species on other media with the goal to understand carbon/nirogen cycling and metabolic capabilities of bacteria prevailing in the reservoirs.
The majority of studies in petroleum microbiology have been conducted using samples of formation water or water–oil mixtures. Only one study reported in the literature used crude oil from petroleum reservoir to analyze microbial communities.45 We isolated DNA from core samples from a petroleum reservoir, and to our knowledge this is the first report of its kind.
The abundance, composition, and diversity of microbial communities within soils are strongly depth dependent.46 In this mesothermic core sample, we elucidated a wide range of bacteria. Alphaproteobacteria dominated the population and were similar to bacterial populations found in seawater and hypersaline lakes. The bacterial diversity is greater as compared to deeper high temperature oil reservoirs under seawater.19,33 However, Alpha-, Beta-, and Gammaproteobacteria are common classes found in both environments. Some of these microbial groups were shown to be exclusive of the biodegraded oil sample, such as the genera Acinetobacter, Bacillus, and Streptococcus. We found some bacteria showing similarity to Streptococcus sp. Presence of other oil degraders in the population as documented by Sette et al (2007) was not evident. In petroleum reservoirs, anaerobic conditions exist, and oil degradation is mainly mediated by methanogenesis.47 Therefore, we believe hydrocarbon utilization is the most important source of nourishment in petroleum deposits, although in some cases the microbes were lithotrophic and survived by utilizing the metal oxides present.
This study also shows that enrichment cultures did not elucidate most of the diversity in the core samples except for some Pseudomonas sp. that were found by both culturing and direct sequencing. However, extraction of DNA from the hard core samples was difficult and required the use of special equipment (Barocycler, PBI). There is the possibility of missing some bacterial species due to differential lysis and PCR biases. However, to decipher the entire microbial diversity of oil reservoirs, this method is necessary along with isolation from crude petroleum and production water. New approaches using high-throughput DNA sequencing will be our next step for obtaining insight into the function and diversity of oil inhabiting bacteria and the pathways for production/degradation of petroleum hydrocarbons.
None.
The author declares no conflict of interest.

©2014 Paul, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.