Journal of
eISSN: 2373-437X


Research Article Volume 6 Issue 6
1Department of Science and Technology, Inter-American University of Puerto Rico, USA
2Department of Biology, University of North Carolina at Charlotte, USA
Correspondence: Richard L. Giles, Department of Science and Technology, Inter-American University of Puerto Rico, Arecibo Campus, P.O. Box 4050, Arecibo, Puerto Rico, 00614 USA, Tel 18284610665
Received: April 27, 2017 | Published: December 26, 2018
Citation: Giles RL, Babilonia LJB, Vargas DG, et al. Bioprospecting filamentous fungi from Puerto Rico for biotechnological applications. J Microbiol Exp. 2018;6(6):250-253. DOI: 10.15406/jmen.2018.06.00224
Novel fungal isolates from Puerto Rico were collected, morphologically identified, axenically cultured, and examined for lignolytic activity. Nine isolates were selected for ABTS differential media assays and rDNA sequencing and identification, five of which exhibited lignolytic activity. The results highlight the need for further conservation and studies on the potential utility of understudied fungi isolated from Puerto Rico. Our study is the first to collect and identify fungi from the island of Puerto Rico with the purpose of examining cultures for potential biotechnological applications. We are the first to demonstrate that wild isolated strains of fungi from Puerto Rico are a potential source of novel fungi to improve current biopulping, bioethanol, and bioremediation technologies.
Keywords: bioprospecting, Puerto Rico, bioremediation, bioethanol, fungi
To date, few Puerto Rican wood decay fungal species and strain isolates have been studied for biotechnology. Wood decay fungi have narrow growth condition requirements such as optimal temperatures for vegetative growth, high humidity for sporocarp formation, lignin type, and substrate specific carbohydrates for metabolic functions.1 Saprobic wood decay fungi are highly host plant specific due to these requirements for growth. Therefore, fungal species habitat range mirrors the geographical distribution of the vascular plant host range and a 6:1 fungus to plant ratio has been observed.2‒4 The biodiversity of fungal species within a geographical region is also reflective of plant diversity although much less study has been devoted to cataloging new fungal taxa.5‒9 While fungal diversity has not been significantly explored in many areas of the world, extrapolations of small habitat fungal inventories provide insight into the potential diversity of understudied areas. Notably, intensive cataloging studies reveal surprisingly high numbers of fungal taxa within small geographic areas. For example, examined leaf litter under five trees in Panama contained 500 different fungal species and a study of palms collected 1,580 Ascomycete species (1,185 were previously undiscovered).10,11
Novel methods utilizing fungal decay mechanisms have been previously examined using as methods for sustainable and economical methods of paper production, chemical remediation, and biofuels produciton.12‒21 However, none of the abundant wild fungal species in Puerto Rico have been isolated for use in this emerging field of biotechnology. Therefore, we propose collecting and characterizing fungal species in the biologically diverse island of Puerto Rico in order to further this area of promising research. To our knowledge, fungi from Puerto Rico have not been isolated and examined for the utility in the varied fields of lignocellulosic degradation technologies. This study combines a fundamental biodiversity study with a screening for an applied microbial degradation ability to progress the study of these unique and potentially useful fungi for biotechnology. To test these hypotheses novel wild isolates fungi from Puerto Rico were identified by morphology and rDNA sequencing, axenically cultured for study, and screened for lignolytic activity utilizing differential media.
Wild fungal isolate collection and identification
Reproductive sporocarps (basidiocarps) were collected in wooded areas in Puerto Rico, USA and returned to the laboratory for inspection. Macroscopic and microscopic characteristics were used to identify the collected specimens to morphological species using standard taxonomic monographs and treatments as previously described.21‒25 Macroscopic characters such as basidiocarp type (e.g. resupinate, applanate), presence/absence of stipe, and margin growth were used for identification. Micro morphology such as context tissue hyphal types (e.g. skeletal, generative) were identified using light microscopy and staining reaction techniques (e.g. Melzner’s reagent).22,23,25 Once identified to morphospecies, sporocarps were split under aseptic conditions and inner tissue samples were inoculated onto non-selective fungal media (20g malt extract, 10mg tetracycline, and 20g agar L-1 in deionized water) and incubated in darkness at 28°C for 7e10 days.26,27 Hyphal colonies were then serially subcultured on new agar plates without antibiotic and maintained as pure cultures at 28°C in darkness on modified malt extract agar (20g malt extract and 20g agar L-1 in deionized water) until use. Axenic fungal isolates that exhibited rapid growth in modified malt extract agar were considered candidates for further study and elucidated using ABTS and rDNA identification.
ABTS Agar plate assays for lignolytic activity
Square agar plugs (1cm2 ) cut from stock malt extract agar plate cultures were used to aseptically inoculate each differential media assay plate. To examine the ability of each fungal isolate to depolymerize ABTS, modified Malt Extract Agar containing ABTS (1 g 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid), 20g malt extract, and 20g agar L-1 in deionized water).28 All treatment combinations and sterile controls were conducted in triplicate. The assay plates were incubated at 28°C in darkness for 21 days to allow adequate time for the fungal mycelia to spread across the surface of each plate, and observed for color changes within the media.28
rDNA Identification
Fungal DNA was extracted from cultured hyphae of isolates using techniques modified from Lõoke et al.29 Approximately 100mg of fungal mycelium was mechanically ground using a rod, mixed with 200mL of 200mM lithium acetate (LiOAc) with 1% sodium dodecyl sulfate (SDS), and then incubated for 15min at 70°C. After incubation, 300mL of 95% ethanol was added and the sample was immediately vortex-mixed (15s) and then pelleted by centrifugation (5min, 15,000g). The supernatant was then replaced with 500mL of 70% ethanol and the pellet was resuspended by vortex mixing (30s) followed by sedimentation by centrifugation as above. The supernatant was then discarded and the pellet was resuspended in 100mL of H2O resulting pellet was resuspended in supernatant was removed and the sample was resuspended in 100mL of H2O. This genomic DNA from each strain was used as template for PCR amplification of the ITS region of the rDNA using primers ITS1-F (sequence: CTTGGTCATTTAGAGGAAGTAA) and ITS4-B (sequence: CAGGAGACTTGTACACGGTCCAG), previously described as specific for amplification of the entire ITS region (ITS1-5.8S-ITS2) of Basidiomycetes fungi.20,30‒32 Individual PCR reactions consisted of 60ng of template DNA, 25µL of GoTaq Green Master Mix (Promega Corp., Madison, WI, USA), 100ng of each primer, and 2.5 mL DMSO with reaction parameters as follows: initial denaturing at 94°C for 2min, 30 cycles of 94°C for 30s, 56°C for 45s, 72°C for 2:15, followed by 72°C for 5min. PCR products were visualized and size checked using gel electrophoresis, and purified using polyethylene glycol (PEG) (Thermo Fisher Scientific Inc., Waltham, MA, USA) and ethanol following Fawcett & Parrow.33 Cleaned products were mixed with appropriate sequencing primers and sequenced in both directions using Applied Biosystems BigDye version 3.1 (GENEWIZ, Inc., South Plainfield, NJ, USA). Resulting sequence assemblies were queried using advanced, nongapped NCBI BLAST search with expectation frequency set to 0.0001 and no filtering for low complexity as previously described.34 The highest BLAST bit score and maximum identity similarity were used to determine closest available taxon to confirm morphological identification.35
Identification of novel fungal isolates and evaluation of lignolytic activity
Wild isolates brought into pure culture for these studies were unambiguously identified to morphospecies by distinct features of their basidiocarp morphology, and identity based on ITS rDNA sequence comparisons confirmed this morphological classification (Table 1).20,22,23,35,36 Among the isolates collected for study, fungi from the taxonomic phyla Ascomycota and Basidiomycota exhibited lignolytic activity in the ABTS media suggesting laccase mediated enzymatic hydrolysis of the compound (Figure 1& 2,Table 1).28 Further analysis of mechanism (e.g. enzymatic) is required to characterize the differences between lignolytic isolates.12,37 The bioprospecting of Puerto Rico’s fungal biodiversity will significantly contribute to future biofuel and bioremediation by making available new species and novel strains for examination. While it is clear that a collection and characterization of fungal taxa of Puerto Rico described in this study is beneficial to understanding forest ecology, it is also important to isolate and culture these organisms for future study. Collection of live cultures will allow future study of decay fungi that may not be obtainable in the future due to habitat loss, providing a snapshot of decay ecology within several study sites. Fungal biodiversity loss due to threat of disease and human encroachment has not been studied in regards to Puerto Rico, however inferences of potential loss can be made because of the known relationship between wood decay fungal diversity and host woody plant diversity.38 Because of this correlation, it is apparent that the decline of tree biodiversity due to disease and loss of habitat strongly suggests that wood decay fungal species may also be declining. Of particular concern is the potential loss of undiscovered and uncharacterized fungal species within the highly specialized forest ecotypes inside Puerto Rico’s forests. Habitat disturbance such as the introduction of non-native disease and human disturbance has been shown to negatively affect the forest biodiversity and ecology of Puerto Rico.39,40 For example, the well known historical example of Cryphonectria parasitica (chestnut blight) which killed native Castanea dentata (American chestnut) tree stands, the dominant hardwood forest species and the major producer of hard mast (e.g. acorns).41 More recent examples of destructive plant pathogens affecting the cove hardwood and spruce-fir forests include dogwood anthracnose and the fir wooly adelgid. Discula destructive (dogwood anthracnose) has killed Cornus florida (flowering dogwood trees) within the Great Smoky Mountain National Park cove forests, significantly changing the understory plant ecology over the last two decades, and possibly affecting nutrient cycling.42,43 These ecological pressures have substantial cascading effects on forest ecology and therefore fungal biodiversity. It is therefore imperative to promote the conservation of these unique fungi and forest ecotypes by careful collection, culturing, and study. Our study is the first to collect and identify fungi from the island of Puerto Rico and examine cultures for potential biotechnological applications.
Taxonomic groups and fungal species |
Extracellular ABTS-oxidizing activity |
|
Basidiomycota |
Trametes cubensis |
+ |
Trametes pubescens |
+ |
|
Ascomycota |
Lasidiplodia pseudotheobromae |
+ |
Aspergillus sp. |
- |
|
Cladosporium sp. |
+ |
|
Fusarium sp. |
- |
|
Curvularia sp. |
+ |
|
Trichoderma sp. |
- |
|
Zygomycota |
Cunninghamella sp. |
- |
Table 1 Identified fungal isolates collected from Puerto Rico and ABTS differential media assay results. Fungi with ABTS depolymerization activity denoted by (+). A (-) indicates no ABTS depolymerization observed.
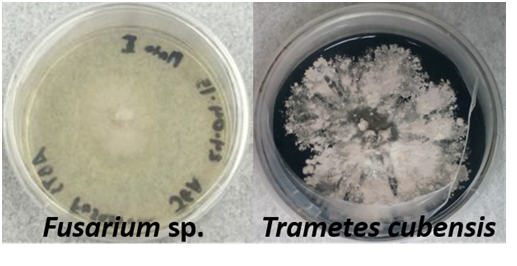
Figure 1 Differential media with modified Malt Extract Agar containing ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) were used to examine the lignolytic capability of fungal isolates from Puerto Rico. The agar turns green due to the oxidation of ABTS. An isolate of Trametes cubensis exhibited significant ABTS oxidation compared to a Fusarium sp. isolate showing no indication of ABTS depolymerization.

Figure 2 Differential media with modified Malt Extract Agar containing ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) were used to examine the lignolytic capability of fungal isolates from Puerto Rico. The agar turns green due to the oxidation of ABTS. An isolate of Curvularia sp. exhibited significant ABTS oxidation compared to an Aspergillus sp. isolate showing no indication of ABTS depolymerization.
Novel fungal isolates from Puerto Rico were identified and examined for lignolytic activity. We are the first to demonstrate that wild isolated strains of fungi from Puerto Rico are a potential source of novel fungi to improve current biopulping and bioremediation technologies. The results highlight the need for further studies on the potential utility of understudied fungi isolated from Puerto Rico.
The authors would like to thank Paul McKenzie for laboratory assistance.
Authors declare that there is no conflict of interest.

©2018 Giles, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.