Journal of
eISSN: 2373-437X


Research Article Volume 6 Issue 5
A.T. Still University, Mesa, AZ, North Carolina State University, Raleigh, NC, USA
Correspondence: Lucia Clontz, Quality Director, Xellia Pharmaceuticals, USA
Received: June 30, 2018 | Published: September 5, 2018
Citation: Clontz L. Biofilm inhibition: the use of a marine alkaloid derivative in the prevention of clinically-relevant biofilms. J Microbiol Exp. 2018;6(5):206-214. DOI: 10.15406/jmen.2018.06.00216
Biofilms are complex and highly resistant microbial communities of sessile cells, which are responsible for many human pathogeneses. Given the risk that biofilms present to public health, this study was performed to contribute to the body of knowledge in the area of infections control by investigating the ability of Agilyte™, a marine alkaloid derivative, to inhibit biofilms of clinical significance and a potential synergistic effect of Agilyte™ and Penicillin G. The bacteria methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, Staphylococcus epidermidis, and Pseudomonas aeruginosa were used in an in-vitro study. Biofilms were established using microtiter plates incubating at 35°C for 24hours, under static and aerobic conditions. To evaluate biofilm inhibition properties and a combined synergistic effect, Agilyte™, with and without antibiotic, was added to bacterial cultures prior to incubation. Biofilm mass was determined using a crystal violet reporter assay and viable cells were determined using the drop-plate method. Independent samples t-test was used to compare biofilm mass from treated samples and positive controls. Statistical significance was set at P<0.05. Mean Log10 Reduction (LR) was calculated for viable plate counts and significant difference was set at >0.3 LR.
The data collected provide evidence that AgilyteTM, at given concentrations and under specific biochemical environments, inhibit biofilms of both gram-positive and gram-negative bacteria. The data suggest that Agilyte™ may inhibit the AI-2 cell signaling mechanism in bacteria. This study did not provide conclusive evidence of a possible synergistic anti-biofouling/antimicrobial effect of Agilyte™ and Penicillin G. However, given that the study yielded some questionable and variable results, data generated using polystyrene microtiter plates should be evaluated with caution and further studies are required to fully evaluate and validate the spectrum of anti-biofouling properties of this chemical compound. The biochemical environment where clinical bacterial biofilms form is an important factor when designing biofilm assays. Therefore, future in-vitrostudies should include accurate and representative models of relevant tissue and medical device surfaces where these microbial communities develop.
Keywords: staphylococcus aureus, microorganisms, biofilms, nosocomial infections, chemical concentration
EPS, extra polymeric substances; VNC, viable but not culturable; MRSA, methicillin-resistant Staphylococcus aureus; CV, crystal violet; LBNS, Luria bertani broth with no salt; NIH, national institutes of health
Biofilms are complex and three-dimensional communities of sessile cells encased in a slimy matrix of extra polymeric substances (EPS) and irreversibly attached to a surface.1 Biofilms are responsible for many human pathogeneses including chronic wounds, device-associated infections, and various nosocomial infections and diseases in patients with compromised immune systems.2 The predominance of biofilms in nature shows that microorganisms have a tendency to form complex microbial communities that are highly resistant to environmental stresses. Understanding how microorganisms develop into biofilm cells and the mechanisms of antimicrobial resistance present in biofilm communities are critical for the development of successful control of bacterial infections, which are a major concern in healthcare and often lead to higher rates of morbidity and mortality in patient populations.3 According to the U.S. National Institutes of Health (NIH), approximately 80% of infections are due to microbial biofilms.4 Biofilms represent a public health risk due to four main reasons. First, it is accepted that microorganisms will develop into biofilms as long as moisture and a substratum are available. Second, when bacteria attach to a surface and develop into a biofilm, these microorganisms become more difficult to remove and kill when compared to planktonic cells or sessile cells that have not yet assumed a biofilm phenotype. Third, biofilm cells can be dispersed by processes such as shedding or by physical forces. Dispersed cells often retain their biofilm phenotype, including antimicrobial resistance and ability to attach to substrata, thus posing a risk of systemic infection. Lastly, biofilms shed toxins to include pyrogenic substances, such as endotoxins produced by gram-negative bacteria.5
Bacteria present in biofilms exhibit an altered phenotype in terms of growth rate and gene transcription. As a result, biofilm cells behave quite differently from their free-floating (planktonic) counterparts. This change in phenotype, combined with the metabolic cooperation among biofilm cells and the unique architecture of a biofilm community, leads to a multi-layer defense mechanism that results in an increased resistance to many chemicals and other types of environmental stresses that are lethal to planktonic cells. Biofilm infections are known for their refractory nature that requires repeated antibiotic treatments.6 In many cases, traditional culture methods used to sample wound sites were unable to recover contaminants, a fact that resulted in the erroneous assumption that chronic wounds were a sterile inflammatory condition. Further studies performed on cultures collected, using molecular biology and microscopic techniques, showed that viable-but-not-culturable (VNC) bacteria were present, metabolically active, and living in a matrix-enclosed community of cells.7 In a study where 50chronic wound specimens were microscopically analyzed, 60% of the samples contained biofilms. Types of bacteria typically found on chronic wound sites are organisms from the genera Staphylococcus, Enter ococcus, Pseudomonas, and the strictly anaerobes Prevotella and Porphorymonas.8
Biofilms are a major deterrent to the use of medical devices due to risk of infection.9 Device-related infections are difficult to treat using conventional antibiotics and they often prove fatal. The mortality rate for bloodstream infections associated with catheters is in the range of 12-25%10 while mortality rate for prosthetic heart valve-related endocarditis is as high as 70%.6 The most common remediation for device-associated infections is removal of the contaminated implant.10 Over the years, many studies have been carried out in the areas of biofilm eradication, remediation, and prevention. However, the realization that biofilms are ubiquitous in nature and extremely difficult to destroy resulted in a shift in paradigm – from microbial eradication and remediation to biofilm prevention strategies. Since then, researchers have worked on novel approaches to create surfaces that prevent microbial adhesion and to develop compounds that prevent or disrupt biofilms. Despite the advances made in the area of biofilm prevention, few studies have been conducted to evaluate the synergistic antimicrobial effect of anti-biofouling compounds and antibiotics. This type of strategy could render bacterial pathogens, to include those that have developed antibiotic resistance, more susceptible to traditional antimicrobial therapies.10
AgilyteTM (Agiles Sciences, Raleigh, NC) is a marine alkaloid derivative of the natural product bromoageliferin produced by reef sponges of the genus Agelas (Family Agelasidae). These organisms produce brominated pyrrole alkaloids, as secondary metabolites, for anti-predatory purposes.11 These chemicals also have the ability to prevent bacterial surface colonization by modulating biofilm formation. Previous studies using a similar bromoageliferin analogue demonstrated enhanced anti-biofilmproperties when used against the gram-positive bacterium Staphylococcus aureus and the gram-negative bacteria Pseudomonas aeruginosa and Acinetobacter baumannii.12 In addition; there was a lack of cytotoxicity detected when AgilyteTM was tested with mammalian cell lines. Given that Agilyte™ is able to maintain and/revert cells to their sensitive planktonic state and it is not cytotoxic, this chemical may act synergistically with conventional antibiotics and serve as an adjuvant for antimicrobial therapies. AgilyteTM is a descriptive term for a family of about 500 non-toxic synthetic organic compounds, each having distinctive and selective abilities to disrupt and disperse bacterial biofilms. The chemical used in this study is AgilyteTM 8000, one of a set of proposed leading anti-biofilm candidates.13
The protocol used to grow biofilms in vitro was an adaptation of the microtiter plate methods described by Ren et al.14 & Pitts et al.15 This test platform is a practical and relevant method to screen various chemical compounds against different types of microbial biofilms. The purpose of this study was to evaluate the ability of Agilyte™ to inhibit biofilms of clinically relevant bacteria. In addition, this study aimed to investigate a potential anti-biofouling synergistic effect between Agilyte™ and antibiotics approved to treat infections.
Selection of bacterial species was based on clinical relevance and their prevalence as isolates from infection sites. The study was conducted using two gram-negative bacteria (Pseudomonas aeruginosaand Escherichia coli) and two gram-positive bacteria (methicillin-resistant Staphylococcus aureus [MRSA] and Staphylococcus epidermidis). The gram-negative bacterium Pseudomonas aeruginosa is an important causative agent of various types of acute and chronic infections that include wound infections at burn sites as well as infections associated with the respiratory system, otorrhea, cornea, and urinary tract system. In cystic fibrosis patients, this organism is the main cause of morbidity and mortality.16 Escherichia coli, a gram-negative enterobacterium, is often associated with urinary tract infections.17 The gram-positive bacterium Staphylococcus epidermidis is a skin commensal and opportunistic pathogen that has become the leading cause of nosocomial and device-related infections.18 The gram-positive bacterium Staphylococcus aureusis a common inhabitant of the human skin flora. This organism is considered a pathogen and it has been associated with many diseases and device-related infections that often lead to bacteremia.19 Given the increased number of cases of septicemia caused by MRSA, and the fact that this bacterium has become a major cause of mortality and morbidity in hospitalized patients2, MRSA was the Staphylococcus aureus type strain used in the study. The antibiotic chose for the study, Penicillin G, is a commonly approved drug for the treatment of bacterial infections humans.
The chosen biofilm assay protocol was an adaptation of the microtiter plate methods described in the literature.14,15 Sterile tissue culture-treated 96-well polystyrene microtiter plates, with clear sides and clear bottoms and with a 300-μl well capacity, were used for the study. To grow biofilms, a 200-microtiter (µl) aliquot of bacterial suspension was added to individual wells and plates incubated for 24hours at 35°C, under stationary and aerobic conditions. Following incubation, spent media and planktonic cells were removed and wells washed, four times, with sterile phosphate Butterfield’s dilution buffer (PBS) to remove loose cells. Biofilms were then quantified using a crystal violet (CV) reporter assay, which uses absorbance (A540) values to represent biofilm mass.15 This was done by staining the sessile cells with 200μl of Gram Crystal Violet (BD Diagnostics) for 10 minutes at ambient temperature (18-25°C). Then, the excess stain was decanted and each well washed, four times, with 200μl of Purified Water (Milli-Qâ, Millipore Corporation). A 200-μl aliquot of Gram decolorizer (BD Diagnostics) was added to each well and the plates incubated for 15minutes at ambient temperature (18-25°C). Blanks were prepared by staining sterile wells following the same procedure used for wells with biofilms. The solubilized stain preparations from each well (including blanks) were transferred into another sterile 96-well microtiter plate and absorbance measured at 540nm using a SpectraMax Plus spectrophotometer (Molecular Devices Inc., Sunnyvale, CA). Raw absorbance values from biofilm wells were corrected by subtracting from the mean blank value.
Bacterial strains
The following bacteria were used in the study: Escherichia coli (ATCC 8539), Staphylococcus epidermidis (ATCC 29886), methicillin-resistant Staphylococcus aureus, MRSA (ATCC BAA-44), and Pseudomonas aeruginosa (PA14).
Media, materials, and reagents
Bacteria were maintained on R2A medium (Difco) under refrigerated conditions (2-8°C). Growth media used for the biofilm assays were BBL tryptic soy broth (TSB), Bacto™ tryptic soy medium with 0.3% glucose broth (TSBG), Luria-Bertani broth (LB), Lennox (BD Diagnostics), Luria-Bertani broth with no salt (LBNS), and M63 medium (1X of 5X stock solution [15g KH2PO4, 35g K2HPO4, 10g (NH4)2SO4, 1 L water]) supplemented with 4% arginine (Sigma). Tryptic soy agar (TSA) and R2A medium (bio Merieux Clinical Diagnostics), and sterile phosphate Butterfield’s dilution buffer (Remel) were used for plate count studies. Agilyte ™ 8000 stock solution was prepared at 1mM concentration in dimethyl sulfoxide (DMSO) and maintained under refrigerated conditions (2-8°C). The studies were performed using concentrations of Agilyte™ ranging from 0.5mM to 500mM. The antibiotic used for the synergistic effect studies was Penicillin G sodium salt (Sigma Aldrich, Catalog # P3032).
Study design
This study was designed to determine the concentrations of Agilyte™ that would inhibit biofilms of clinically relevant bacteria. The minimum biofilm inhibitory concentration (MBIC) values were determined and used to evaluate a potential anti-biofouling synergistic effect of Agilyte™ and Penicillin G. Biofilm Inhibition Determination. To study the anti-biofilm properties of AgilyteTM, this chemical was added to an overnight bacterial culture adjusted to an optical density (OD) 600 of 0.01- 0.03. TSB and TSBG were used in studies with MRSA and S. epidermidis. TSB and LB were used in studies with E. coli. TSB, LB, LBNS, and M63 with arginine were used in studies with PA14. Various concentrations of AgilyteTM were evaluated to determine the MBIC for a given bacterial species. Bacterial suspensions without AgilyteTM were treated as positive controls. A minimum of five replicates were prepared for each sample treatment and positive control. Biofilms were established in microtiter plate wells and quantified using a CV reporter assay, as described above. Percent biofilm inhibition was determined by comparing the mean absorbance value for biofilms formed in the presence of AgilyteTM to the mean absorbance value for positive control biofilms.
Determination of synergistic biofilm inhibition effect
To study a potential synergistic anti-biofouling effect, Agilyte™ at the MBIC level and Penicillin G at a non-inhibitory concentration for a given species were added to overnight bacterial cultures, which had been adjusted to an OD600 of 0.01-0.03. TSBG and Penicillin G at 25Μm20 were used for studies with MRSA. TSBG and Penicillin G at 0.06μM21 were used with studies with S. epidermidis. Controls were prepared by growing biofilms in the presence of Penicillin G only and in the presence of AgilyteTM only. Positive controls were prepared by growing biofilms in the absence of both AgilyteTM and Penicillin G. A minimum of five replicates were prepared for each sample treatment and controls. Biofilms were established in microtiter plate wells and quantified using a CV reporter assay, as described above. Percent biofilm inhibition was determined by comparing the mean absorbance value for biofilms formed in the presence of Agilyte TM, antibiotic, and AgilyteTM with antibiotic to the mean absorbance value for positive control biofilms.
For some of the studies, viability of biofilm cells was determined using the drop plate method.22 Following incubation at 35°C for 24-hours, under stationary and aerobic conditions, spent media and planktonic cells were removed and wells washed, four times, with sterile PBS to remove loose cells. Then, 200-ml aliquots of sterile PBS were added to replicate wells, which were scraped with sterile plastic applicator sticks (Fischer Scientific Co., catalog # 23-400-122) to dislodge the sessile cells. The buffer-cell suspension was aspirated and added to 4.6 ml of sterile PBS contained in a test tube. Another 200-ml aliquot of sterile PBS was added to the same wells, the scraping procedure repeated, and the buffer-cell suspension aspirated and added to the same test tube with sterile PBS for a final volume of 5ml. The buffer-cell preparation was vortexed for 30seconds and sonicated for 30seconds using the Aquasonic Model 50D sonicator (VWR Scientific, West Chester, PA). This procedure for biofilm disaggregation 23 was repeated twice and the cell suspensions serially diluted and drop plated22 using R2A medium or TSA. Plates were incubated at 35°C for 17 to 20hours. Following incubation, colonies recovered were enumerated using the Quebec colony counter (Reichert Analytical Instrument, Depew, NY). An appropriate dilution factor was used to determine the number of viable cells (biofilm mass) per well.
Statistical analysis
For experiments using the CV reporter assay, each raw absorbance value was corrected by subtracting the mean absorbance for blank wells prior to statistical analysis. For the biofilm inhibition method, an independent samples t-test was performed for comparisons of biofilm mass for the treated samples (mean of corrected absorbance values) and for the positive controls (mean of corrected absorbance values). Differences were considered significant for p values of < 0.05, two-tailed. A measure of efficacy called Percent Biofilm Inhibition was calculated for treated samples that were statistically different from test controls. (Equation 1) Let A denote the corrected mean absorbance value for the positive control and B denote the corrected mean absorbance value for the treated sample.
Percent Biofilm Inhibition = (A - B) / A x 100 [Eq. 1]
For each set of experiments, the standard deviation (SD) of replicate measurements was also calculated to establish between-well variability. Then, the mean and 95% confidence intervals (CI) for the replicate Percent Biofilm Inhibition values were calculated. Data were reported as Percent Biofilm Inhibition ± CI. All statistical analyses were performed using Microsoft® Excel 2007. For experiments to determine viability of biofilm cells, the number of colony forming units (viable cell density) was determined for replicate wells. Then, the average viable cell density was calculated and log10 transformed. Log reduction (LR) was calculated as the log10 density for treated wells subtracted from the log10 density for control wells. Differences were considered significant for LR values of > 0.3. 24 Data were reported as Mean Biofilm Mass (log10 CFU/well) and Log10 Reduction.
Effect of agilyte™ on MRSA biofilm inhibition
Agilyte™ was tested for its impact on biofilm formation of MRSA (ATCC BAA-44) grown in TSB and TSBG media. Colorimetric assays of stained biofilm cells demonstrated that Agilyte™ prevents biofilm formation of MRSA growing at 35°C for 24hours, under aerobic and static conditions. When MRSA was grown in TSB, biofilm inhibition was variable and there was no statistically significant difference (p>0.05) between biofilm mass formed in the presence and absence of Agilyte™. However, statistically significant changes in bacterial behavior were observed (p<0.05) when MRSA was grown in TSBG. After a 24-hour incubation period, Agilyte™ at various concentrations inhibited biofilm formation. Mean biofilm inhibition values for ten replicate samples (two separate experiments of five replicates each) treated with Agilyte™ at concentrations of 100µM, 150µM, and 200µM were 37.9%, 92.6%, and 80.3%, respectively (Table 1). Results obtained with Agilyte™ tested at concentrations lower than 200µM were more variable and for many experiments, there was no statistically significant difference (p > 0.05) between samples treated with Agilyte™ and test controls (Table 1). Therefore, Agilyte™ at 200µM was selected as the MBIC and used in studies to evaluate the synergistic effect of Agilyte™ and Penicillin G against MRSA.
|
Agilyte™ Concentration (µM) |
|||||||||
|
Experiment |
No. Replicates |
50 |
60 |
80 |
100 |
120 |
150 |
200 |
Growth medium |
|
1 |
5 |
NC |
NCN |
CNC |
− |
− |
TSB |
||
|
2 |
5 |
− |
− |
− |
NC |
NCN |
CNC |
TSB |
|
|
3 |
5 |
− |
− |
64.6 |
63.2 |
− |
73.6 |
78.3 |
TSBG |
|
4 |
5 |
− |
− |
NC |
12.6 |
− |
111.6 |
− |
TSBG |
|
5 |
5 |
29.5 |
− |
NC |
NC |
− |
NC |
82.2 |
TSBG |
|
Mean CI |
|
|
|
|
37.9 |
|
93 |
80.3 |
|
|
|
|
|
|
|
± 35.1 |
|
±26.3 |
±2.7 |
|
Table 1 Test Results for MRSA–Mean Percent Biofilm Inhibition in the Presence of Agilyte™
Synergistic effect of agilyte™ and penicillin G on MRSA biofilm inhibition
To study the synergistic effect of Agilyte™ and Penicillin G on biofilm formation, MRSA (ATCC BAA-44) was grown in TSBG at 35°C for 24 hours, under aerobic and static conditions, and in the presence of Agilyte™ at 200µM and Penicillin G at 25µM (maximum non-inhibitory concentration). Colorimetric assays of stained biofilm cells from 12 replicate samples demonstrated a potential synergistic biofilm inhibition effect of Agilyte™ and Penicillin G under the given test conditions. As expected, when MRSA was grown in the presence of 25µM Penicillin G only, there was no statistically significant difference (p = 0.67) between biofilm mass from treated samples and test positive controls (biofilms grown in TSBG only). When MRSA was grown in the presence of 200µM Agilyte™ only, unexpectedly, no statistically significant difference (p = 0.73) between biofilm mass from treated samples and test positive controls was detected. However, for the wells that contained both 200µM Agilyte™ and 25µM Penicillin G, biofilm formation was inhibited by 54.4% (p = 0.02). Due to the variability in assay results for this experiment, this test is considered inconclusive since biofilm inhibition could have been due solely to the presence of Agilyte™ in the test sample.
Note. Dashes indicate chemical concentration not tested. NC = no changes in biofilm mass when compared to controls (p > 0.05).
For comparative purposes, viable plate counts were performed. For each replicate testing, three wells from each sample treatment were selected. Biofilm mass collected was serial diluted in PBS and plated with R2A medium. Test plates incubated at 35°C for 16-20 hours. For Replicate 1, plate count results did not show changes in viability of biofilm cells for treated and untreated samples. Serial dilutions for Replicate 2 samples were refrigerated (2-8°C) for 18 hours prior to plating. As expected, there was no viability loss for biofilm cells from test positive control and Agilyte™ sample preparations. However, refrigeration resulted in loss of viability for biofilm cells in the presence of 25µM Penicillin G (1.3 log10 reduction) and a potential antimicrobial synergistic effect (1.9 log10 reduction) between 200µM Agilyte™ and 25µM Penicillin G (Table 2).The 0.6 difference in log reduction between samples treated with Penicillin G and samples treated with Penicillin G and Agilyte™ can be considered greater than normal variability in microbial count (i.e., >0.3log), and therefore a possible indication of enhanced biofilm inhibition effect.
|
Sample |
Mean Biofilm Massa(log10 CFU/well) |
Mean Biofilm Massb(log10 CFU/well) |
Log10Variabilityc |
|
Positive control |
6.3 |
6.4 |
− |
|
Agilyte™ (200µM) |
6.7 |
6.6 |
+0.2 |
|
Penicillin G (25µM) |
6.5 |
5.1 |
-1.3 |
|
Agilyte™ (200µM)/ |
6.5 |
4.5 |
-1.9 |
|
Penicillin G (25µM) |
|
|
|
Note. CFU = colony forming unit. Mean results based on triplicate samples. Dash indicates data not applicable
Effect of agilyte™ on S. epidermidis biofilm inhibition
Agilyte™ was tested for its impact on biofilm formation of S. epidermidis (ATCC 29886) grown in TSB and TSBG. Colorimetric assays of stained biofilm cells demonstrated that Agilyte™ inhibits biofilm formation of S. epidermidis growing at 35°C for 24 hours, under aerobic and static conditions. Biofilm inhibition was similar but slightly greater in TSB when compared to TSBG. Mean Biofilm inhibition values for five replicate samples in TSB treated with Agilyte™ at concentrations of 80µM, 100µM, and 120µM were 84.5%, 89.7%, and 87.6%, respectively (Table 3). Mean biofilm inhibition values for 10 replicate samples (two separate experiments of five replicates each) treated with Agilyte™ at concentrations of 80µM and 100µM were 74.3% and 78.3%, respectively (Table 3). The biofilm inhibition value based on five replicate samples treated with Agilyte™ at a concentration of 150µM in TSBG was 70.3%. Some replicate results obtained with Agilyte™ tested at concentrations equal to 60µM in TSB, and 150µM and 200µM in TSBG were not statistically significant different (p > 0.05) from test positive controls. Based on data collected, Agilyte™ at 100µM was selected as the MBIC and used in studies to evaluate the synergistic effect of Agilyte™ and Penicillin G against S. epidermidis.
|
|
|
|
Agilyte™ concentration (µM) |
|
|
|
||
|---|---|---|---|---|---|---|---|---|
|
Experiment |
No. Replicates |
60 |
80 |
100 |
120 |
150 |
200 |
Growth Medium |
|
1 |
5 |
NC |
84.5 |
89.7 |
87.6 |
− |
− |
TSB |
|
2 |
5 |
− |
50.5 |
50.5 |
− |
NC |
NC |
TSBG |
|
3 |
5 |
− |
87.8 |
94.6 |
− |
70.3 |
− |
TSBG |
|
Mean (TSBG only) |
69.2 |
72.6 |
||||||
|
Mean (TSBG and TSB) |
74.3 |
78.3 |
||||||
|
CI (TSBG/TSB) |
±23.3 |
±27.4 |
|
|
|
|
|
|
Table 3 Test Results for S. epidermidis – Mean Percent Biofilm Inhibition in Agilyte™
Note. Dashes indicate chemical concentration not tested. NC = no changes in biofilm mass when compared to controls (p > 0.05).
Synergistic effect of agilyte™ and penicillin G on S. epidermidis biofilm inhibition
To study the synergistic effect of Agilyte™ and Penicillin G on biofilm formation, S. epidermidis (ATCC 29886) was grown in TSBG at 35°C for 24 hours, under aerobic and static conditions, and in the presence of 100µM Agilyte™ and 0.06µM Penicillin G (maximum non-inhibitory concentration). Colorimetric assays of stained biofilm cells from 12 replicate samples did not show a synergistic effect of Agilyte™ and Penicillin G.As expected, mean colorimetric assay values of stained biofilm cells showed no statistically significant difference (p = 0.69) between biofilm mass formed in the presence of 0.06µM Penicillin G and biofilm mass from the test positive control. Also as expected, a mean 65.5% biofilm inhibition in the presence of 100µM Agilyte™(p < 0.001) was observed. In addition, in the presence of both 100µM Agilyte™ and 0.06µM Penicillin G (p < 0.001) a mean 64.4% biofilm inhibition was observed. No additional biofilm inhibition was evident as a result of a possible synergistic effect between Agilyte™ and Penicillin G at the concentrations tested. Figure 1 shows a summary of results obtained. No viable plate counts were performed for comparative purposes.
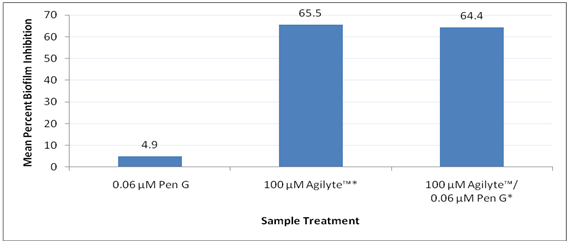
Figure 1 Synergistic inhibition effect of Agilyte™ and Penicillin G on S. epidermidis biofilms. Treated samples compared to test positive controls. Mean percent biofilm inhibition based on 12 replicate samples. Note. *p < 0.001.
Effect of agilyte™ on E. coli biofilm inhibition
Agilyte™ was tested for its impact on biofilm formation of E. coli (ATCC 8739) grown in TSB and LB media. Colorimetric assays of stained biofilm cells were used to quantify biofilm mass. Two separate experiments performed showed that when E. coli was grown in TSB at 35°C for 24 hours, under aerobic and static conditions, biofilm growth increased with increased concentration of Agilyte™ (Figure 2). However, when E. coli was grown in LB at 35°C for 24 hours, under aerobic and static conditions, biofilm mass decreased as concentration of Agilyte™ increased (Figure 3). Mean percent E. coli biofilm inhibition values for five replicate samples treated with Agilyte™ were as follows: 30µM (37.8%, p = 0.04), 50µM (61.2%, p < 0.001), 80µM (86.6%, p = 0.15; 63.8%, p = <0.001), 100µM (85.3%, p = 0.15; 87.2%, p = <0.001), 150µM (84.9%, p = 0.15), and 200µM (75.6%, p = 0.20; 79.3%, p = < 0.001)
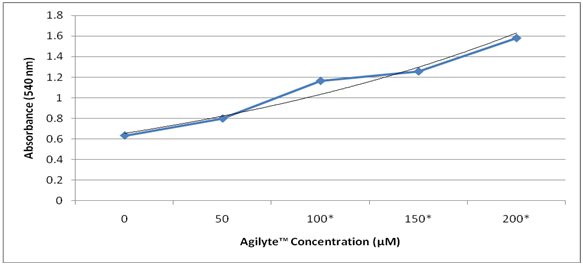
Figure 2 Effect of Agilyte™ on E. coli biofilms grown in TSB at 35°C for 24hours. Experiment 2. Mean absorbance values based on five replicate samples. Note. *p < 0.05.

Figure 3 Effect of Agilyte™ on inhibition of E. coli biofilms grown in LB at 35°C for 24 hours. Mean results based on five replicate samples. Treated samples compared to test positive controls. Note. * p < 0.05, ** p < 0.001.
Effect of agilyte™ on pseudomonas aeruginosa biofilm inhibition
Agilyte™ was tested for its impact on biofilm formation of P. aeruginosa (PA14) grown in various types of media including TSB, LB, LBNS, and M63 with arginine. Colorimetric assays of stained biofilm cells were used to quantify biofilm mass. When PA14 was grown in TSB medium at 35°C for 24 hours, under aerobic and static conditions, Agilyte™ at the concentrations tested enhanced biofilm formation (Figure 4). A similar bacterial behavior was observed when PA14 was added to LB medium spiked with Agilyte™ and allowed to grow at 35°C for 24 hours, under aerobic and static conditions. However, although biofilm enhancement was observed across a wide chemical concentration range (0.5µM to 500µM), at low (0.5µM, 5µM, and 10µM) and high (500µM) concentrations, biofilm formation was more pronounced as compared to biofilms formed for samples treated with 50µM, 100µM, and 200µM of Agilyte™ (Figure 5) (Table 4). Bacterial behavior in LBNS treated with Agilyte™ was variable and inconsistent. For example, in one study, 100 µM of Agilyte™ yielded a 26.4% (p = 0.03) mean biofilm inhibition while in a replicate study, the same concentration of Agilyte™ led to a 67.2% (p < 0.001) mean increase in biofilm formation (Figure 6). Percent values were calculated in comparison to test positive controls.
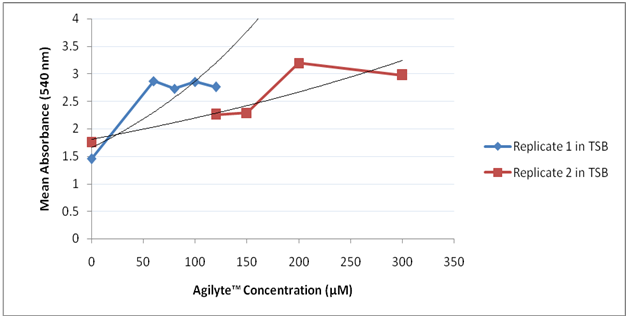
Figure 4 Effect of Agilyte™ on PA14 biofilms grown in TSB at 35°C for 24 hours. Mean absorbance based on five replicate samples.

Figure 5 Effect of Agilyte™ on PA14 biofilms grown in LB at 35°C for 24 hours. Mean absorbance based on seven replicate samples.

Figure 6 Effect of Agilyte™ on PA14 biofilms grown in LBNS. Mean absorbance values for Replicate 1 study based on five replicate samples. Mean absorbance values for Replicate 2 study based on six replicate samples. Note. * p < 0.05.
|
Sample Agilyte™ concentration (µM) |
||||||||
|---|---|---|---|---|---|---|---|---|
|
|
0 (PC) |
0.5 |
5 |
10 |
50 |
100 |
200 |
500 |
|
Mean Biofilm Massa(A540) |
0.73 |
0.899 |
0.989 |
1.077 |
0.866 |
0.805 |
0.751 |
1.046 |
|
SD |
0.08 |
0.899 |
0.28 |
0.33 |
0.19 |
0.07 |
0.15 |
0.05 |
|
pb |
− |
0.06 |
0.04 |
0.02 |
0.11 |
0.08 |
0.75 |
< 0.001 |
A low-nutrient medium, M63 with arginine, was then used to grow PA14 biofilms. Following a 24-hour incubation period at 35°C, under aerobic and static conditions, there was no turbidity in the medium contained in the inoculated wells. Under similar incubation conditions, PA14 preparations in TSB, LB, and LBNS media produced turbidity with planktonic, sessile cells, and pellicle formation. To increase biofilm mass for optimum quantitation, microtiter plates with PA14 in M63 medium with arginine continued incubation for a total of six days. Then, plates were removed from the incubator and observed for biofilm formation. There was still little to no visual turbidity as a result of planktonic cell growth. However, biofilms were present on the side and bottom of the wells.
Colorimetric assays of stained biofilm cells demonstrated that Agilyte™ prevents biofilm formation of PA14 grown in M63 medium with arginine. However, there was no direct correlation between biofilm inhibition and concentration of Agilyte™. For samples treated with 0.5µM, 5µM, 10µM, and 500µM of Agilyte™, mean biofilm inhibition, in comparison to test positive controls, was 31.2% (p < 0.001), 29.4% (p < 0.01), 47.0% (p < 0.001), and 86.0% (p < 0.001), respectively. For samples treated with 50µM of Agilyte™, there was no statistically significant difference between biofilm mass for treated samples and test positive controls (p = 0.07) and for samples treated with 100µM and 200µM of Agilyte™, biofilm mass increased, in comparison to test positive controls, by 16.3% (p = 0.04) and 64.7% (p < 0.001), respectively (Figure 7).
In this study, we examined the effect of AgilyteTM on inhibiting biofilms of clinically relevant bacteria. Our findings provide evidence that Agilyte™ can prevent biofilm formation of MRSA, E. coli, S. epidermidis, and P. aeruginosa (PA14) under specific conditions. However, data collected were variable and confirmed that bacterial behavior and biofilm formation are dependent on available nutrients and environmental conditions. Therefore, anti-biofilm activity appears to be dependent on bacterial behavior in specific biochemical environments rather than a generic phenomenon correlated to concentrations of a given chemical, such as AgilyteTM. In addition, AgilyteTM showed variable and, sometimes, agonistic effects on biofilm formation when high nutrient media were used to grow gram-positive and gram-negative bacteria. For example, Agilyte™ inhibited MRSA biofilms grown in TSBG and S. epidermidis biofilms grown in TSB and TSBG but enhanced E. coli and PA14 biofilms grown in TSB. AgilyteTM also inhibited E. coli biofilms but not PA14 biofilms, both grown in LB medium. Our data showed that Agilyte™ only inhibited PA14 biofilms grown in a very-low nutrient medium (M63) supplemented with arginine.
Colorimetric assays could not provide conclusive evidence of a combined synergistic inhibitory effect of 200 µM Agilyte™ and 25µM Penicillin G on MRSA biofilms. Despite a difference in biofilm mass determined using a CV reporter assay, plate counts did not show a difference in cell viability between treated samples and positive controls, and between biofilms treated with Penicillin G only and those treated with both Agilyte™ and Penicillin G. These data show that biofilm mass determined by colorimetric methods does not always correlate with cell viability. A duplicate set of dilutions with biofilm cells recovered from the microtiter plate wells was refrigerated (2-8°C) for 18 hours prior to plating with R2A medium. As expected, there were no changes in count (within 0.3log) for the positive control and samples treated with Agilyte™, thus confirming the stability of microbial populations under short-term refrigeration. However, cell viability in the presence of 25µM of Penicillin G and in the presence of both 25µM of Penicillin G and 200µM Agilyte™ was reduced by 1.3log10 and 1.9 log10, respectively. These data show not only increased antibiotic activity, due to a slowdown in cell metabolism, but also a synergistic antimicrobial effect between Agilyte™ and Penicillin G under cold conditions. Thus, these results open the possibility of using Agilyte™ as an adjuvant for topical antimicrobial therapies (e.g., skin wounds) used in combination with cold therapies, which already have proven benefits in reducing inflammation. No synergistic biofilm inhibition effect of Agilyte™ and Penicillin G, at the concentrations tested, was observed for S. epidermidis. A mean 65.5% biofilm inhibition in the presence of 100µM Agilyte™ was observed in comparison to a mean 64.4% inhibition for biofilms treated with both 100µM Agilyte™ and 0.06µM Penicillin G. It is worth noting that this experiment was conducted using TSBG and not TSB, which yielded a greater biofilm inhibitory effect when tested with Agilyte™ alone. Perhaps results would have been different using an alternate growth medium.
Quorum sensing (QS) plays a key role in the initial stages of biofilm formation and in biofilm dispersion. This phenomenon is a type of cell to cell signaling mechanism that enables a bacterium to regulate gene expression in response to cell population density and facilitates an organism’s adaptation to changing environmental conditions. QS depends on the production of small diffusible signal molecules called auto inducers (AI) that fall into three chemical families: acylated homoserine lactones (AI-1) found in gram-negative bacteria, oligopeptides found in gram-positive bacteria, and a system designated AI-2 that is mediated by a highly conserved gene (luxS) and is used by both gram-negative and gram-positive bacteria. Besides QS, AI-2 controls many other bacterial traits and its target receptor is species-dependent. Because of its unique characteristics, AI-2 is believed to be an interspecies communication system.25
Given that AgilyteTM affects biofilm formation of both gram-positive and gram-negative bacteria, it is hypothesized that AgilyteTM might play a role in regulating the AI-2 cell signaling mechanism. Furan one compounds, also derived from marine organisms, are known to interfere with the AI-2 bacterial communication system.26 This hypothesis may be supported by significant differences in the effect that Agilyte™ has on biofilm formation for gram-negative and gram-positive bacteria growing in TSB and TSBG, which contain glucose. Studies have provided evidence that AI-2 is produced in the presence of preferred carbohydrates and metabolized when primary nutrients are depleted27 and that AI-2 uptake is necessary for biofilm formation28 Among carbohydrates that can be metabolized, glucose is preferred by many bacteria including Staphylococcus spp.29 Therefore, it is expected that in TSB/TSBG, S. epidermidis and MRSA would produce and internalize AI-2 as the primary cell signaling mechanism. Data collected suggest that MRSA may require a higher concentration of glucose, which is present in TSBG when compared to TSB to utilize the AI-2 as the main cell-to-cell signaling mechanism. E. coli grown in LB medium releases and accumulates AI-2. However, when E. coli is grown in the presence of glucose, AI-2 cannot be internalized, due to catabolite repression, and accumulates in the medium.30 Therefore, it is suggested that in LB medium, E. coli uses AI-2 as a cell signaling system for biofilm formation but in TSB, the primary AI-1 system is used. The hypothesis that Agilyte™ regulates the AI-2 cell signaling system explains why this chemical inhibited biofilm formation of S. epidermidis and MRSA growing in TSB/TSBG and of E. coli growing in LB but could not inhibit biofilm formation of E. coli growing in TSB. Pseudomonas aeruginosa uses the two major and distinct QS regulatory systems (lasR-I and rhlR-I), which work in concert and are controlled via a hierarchal process .31 In addition, this bacterium has a separate cell signaling system called the Pseudomonas quinolone signal, which is induced by lasR-I and which induces the RhlR-I system. P. aeruginosa does not contain the luxS gene or generate AI-2 signaling molecules. It does, however, express the receptor complex of the AI-2 two-component QS system. Given that P. aeruginosa does not produce AI-2, the lack of biofilm inhibition when PA14 was cultured in TSB, LB, and LBNS could be explained by the hypothesis that Agilyte™ regulates the AI-2 cell signaling system. However, Agilyte™ successfully inhibited PA14 biofilm formation, at certain concentrations, when this bacterium was grown in low-nutrient medium using arginine as the sole nitrogen and carbon source. To explain this result, two hypotheses are proposed. First, it is possible that PA14, in a low-nutrient chemical environment, produces a QS cell signaling molecule, homologous to the one from the AI-2cell signaling system, which is detected by the AI-2 receptor. Thus, it is possible that PA14 uses the AI-2 system not only to detect signals from other bacterial populations but also as an alternative cell signaling detector that is part of its complex QS system. If this hypothesis holds true, it is possible that Agilyte™ inhibits biofilm formation by blocking the AI-2 receptor rather than down regulating transcription of the luxS gene. The second hypothesis is a possible synergistic anti-biofilm effect between arginine and Agilyte™ for P. aeruginosa biofilms. A study performed demonstrated that arginine can increase the susceptibility of P. aeruginosa to two classes of antimicrobials in mature biofilms.32 Therefore, the results obtained with PA14 grown in a medium containing arginine could support the possibility of such synergistic effect.
The ability of bacteria to form biofilms is recognized as important virulence and antimicrobial resistant factors. The results presented here demonstrate that Agilyte™, at given concentrations and under specific biochemical environments, inhibits biofilms of both gram-positive and gram-negative bacteria. The data suggest that Agilyte™ may inhibit the AI-2 cell signaling mechanism in bacteria. This study did not provide conclusive evidence of a possible synergistic anti-biofouling/antimicrobial effect of Agilyte™ and Penicillin G. However, given that the study yielded some questionable and variable results, data generated using polystyrene microtiter plates should be evaluated with caution and further studies are required to fully evaluate and validate the spectrum of anti-biofouling properties of this chemical compound. The biochemical environment where clinical bacterial biofilms form is an important factor when designing biofilm assays. Therefore, future in-vitrostudies should include accurate and representative models of relevant tissue and medical device surfaces where these microbial communities develop. Consideration should be given to experiments with simulated body fluid (SBF) and simulated wound models. Only through such realistic ex-vivo models will one be able to fully explore the effect of anti-biofilm chemicals, such as Agilyte™, on inhibiting and/or disrupting biofilms of clinically relevant bacteria as well as their potential use as an adjuvant in antimicrobial therapies.
This study was performed as an Applied Research project in Partial Fulfillment of the Requirements for the Doctor of Health Sciences degree from A.T. Still University (Mesa, AR) and approved by Jeffrey L. Alexander, PhD and Helen Ewing, RN, DHSc (A.T. Still University, Mesa, AR) and John Cavanagh, PhD (Department of Molecular and Structural Biochemistry, at North Carolina State University, Raleigh, NC). The Study was supported by Diosynth Biotechnology (Morrisville, NC). I am grateful to Dr. Christian Melander (Department of Chemistry, at North Carolina State University, Raleigh, NC) for his technical support and for generously providing the chemical Agilyte™ 8000, Luria-Bertani broth with no salt (LBNS), and the bacteria Staphylococcus epidermidis ATCC 29886, methicillin-resistant Staphylococcus aureus (ATCC BAA-44), and Pseudomonas aeruginosa PA14. I am also grateful to Dr. John Cavanagh for his support and guidance as the External Committee Member for my Applied Research Project. Finally, I would like to thank DB Diagnostics (MD, USA) for providing the Luria-Bertani broth used in the studies.
The author declares that there is no conflict of interest.

©2018 Clontz. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.