Journal of
eISSN: 2373-437X


Research Article Volume 1 Issue 4
1The East Central and Southern Africa Health Community, Tanzania
2College of Health Sciences, Jomo Kenyatta University of Agriculture and Technology (JKUAT), Kenya
3Centre for Public Health Research, Kenya Medical and Research Institute (CPHR, KEMRI), Kenya
4Centre for Virus Research, Kenya Medical and Research Institute (CVR, KEMRI), Kenya
Correspondence: Martin Matu, The East Central and Southern Africa Health Community (ECSA-HC), P. O. Box 1009, Arusha, Tanzania, College of Health Sciences, Jomo Kenyatta University of Agriculture and Technology (JKUAT), P. O. Box 62000-00200, Nairobi, Kenya
Received: June 20, 2014 | Published: July 19, 2014
Citation: Matu M, Kikuvi G, Wanzala P, et al. Aetiology of acute respiratory infections in children under five years in Nakuru, Kenya. J Microbiol Exp. 2014;1(4):122-129. DOI: 10.15406/jmen.2014.01.00021
Introduction: Acute Respiratory Infections (ARI) is the most common causes of both illness and mortality.
Objectives: This study aimed to identify the aetiology of acute respiratory illnesses in subjects from selected health facilities in Nakuru Country in Kenya.
Method: This was a case control study which recruited parents or guardians with children less than 5 years of age who visited the selected health facilities with suspected ARI. Controls were matched for age and sex. Throat and nasopharyngeal swabs were collected from the children to isolation and detection of bacterial and viral agents respectively. A questionnaire was used to obtain socio-demographic information from the study subjects. Logistic regression analysis was used to identify the factors associated with ARI agents.
Results: Bacteria were isolated in 24% of sampled patients with Streptococcus pyogenes and Streptococcus viridans, being the most predominant. At least one ARI respiratory associated virus in 44.9% of the specimen collected from the children with Influenza A (20.5%), RSV (16.7%) and Influenza B (10.3%) being more common. Mixed infections were present in 29.4% of the sampled children. The clinical predictors of bacterial isolation were fever, high respiratory rate, cough and stridor while high respiratory rate and cough were more associated with viral detection.
Conclusion: The study revealed that influenza viruses and Streptococcus pyogenes are the major viral and bacterial etiologies of ARI, respectively, in this study population. Mixed infections are also common among the study subjects.
Keywords: acute respiratory infections, influenza viruses, Streptococcus pyogenes, aetiological agents, mixed infections
ARI, acute respiratory infections; URTIs, upper respiratory tract infections; IVs, influenza viruses; RSV, respiratory syncytial virus; PIVs, parainfluenza viruses; KIPyV, KI polyomaviruses; WUPyV, WU polyomaviruses; hMPV, human metapneumovirus; KEMRI, Kenya medical research institute; GC, gonococci; WHO, world health organization; CDC, centers for disease control; cDNA, complimentary DNA; RT-PCR, reverse transcriptase PCR; SSC, scientific steering committee; ERC, ethical review committee; NK, natural killer.
Acute respiratory tract infections are the most common illnesses in all individuals, regardless of age or gender. Of all acute illnesses, respiratory conditions are the most common, generally occurring twice as frequently as the next most common condition. ARI contributes 2% to 4% of deaths in children <5 years of age in developed countries, and 19% to 21% in the Eastern Mediterranean, Africa, and South East Asia regions.1
Acute respiratory infections are caused mainly by viruses but bacterial aetiologies have been confirmed. A Dutch case-control study of general practice patients found that viruses accounted for 58% of acute respiratory tract infections, bacteria (Group A streptococcus) 11% and 3% of patients had mixed bacterial and viral infections.2 Various viral and bacterial strains have been associated with ARI as described below. There are a number of viruses that are considered primarily as pathogens of the respiratory tract. Over 200 different viruses have been isolated in patients with upper respiratory tract infections (URTIs). Until recently when molecular techniques have been developed for detection and confirmation of various pathogens; viral pathogens have historically been detected using cell culture, antigen detection, or serological diagnostic panels. This group includes the influenza viruses (IVs), Respiratory Syncytial virus (RSV), and the Parainfluenza viruses (PIVs) types I, II and III. Adenoviruses cause respiratory disease, but specific serotypes can affect other organ systems and produce nonspecific illness, such as fever with no clear focus. There is a second line of agents, including rhinoviruses and other picornaviruses, and coronaviruses, that are relatively common, but have previously been thought of as typically associated with milder upper respiratory tract illness.3 Other viruses that have been associated with respiratory infections are Mimivirus and Mossman virus,4 Reovirus, Melaka virus,5,6 the Herpesviruses family, including Human herpes virus type 6, Cytomegalovirus, Varicella‐zoster virus, Epstein‐Barr virus, and the Herpes simplex viruses, particularly in the immunocompromised,7 Human metapneumovirus (hMPV),8 ARS‐associated coronavirus,9 Human bocavirus10 and two Human polyomaviruses: KI (KIPyV) 24 and WU virus (WUPyV).11 These viruses comprise the core group of common childhood respiratory pathogens, as well as those that may only occasionally cause illness.
Up to 15% of acute pharyngitis cases are of bacterial aetiology, commonly Group A Streptococcus in Streptococcal pharyngitis.12 Major LRTIs are associated with various bacterial aetiologies such as Streptococcus pneumoniae, Mycoplasma pneumoniae, Chlamydia spp, Pseudomonas spp., Escherichia coli, and Haemophilus influenzae causing pneumonia. There are few data on the causes of neonatal pneumonia in developing countries, but studies of neonatal sepsis suggest that these include gram-negative enteric organisms, particularly Klebsiella spp., and Gram-positive organisms, mainly Streptococcus pneumoniae, Group B Streptococcus and S. aureus.13 In a study conducted in Hongkong to investigate the bacterial etiology of ARI, Mycoplasma spp., was found to contribute to 3% of the positive cases.14 Vong et al.,15 in a recent study on viral and bacterial aetiologies of community acquired acute lower respiratory infections found that bacteria commonly associated LRTIs in under 5 year-old children were Burkholderia pseudomallei, Streptococcus pneumoniae, Haemophilus influenzae, Klebsiella pneumoniae and Mycobacterium tuberculosis. This study was done to determine the aetiology of ARI to provide information that could aid in the control and management of ARI in the region.
Study design and site
This was a case control study conducted in Nakuru Central District with in a rural and an urban health facility. The study was conducted between the months of April to November 2014.
Study population
The study enrolled caretakers and their children less than 5 years diagnosed with ARI and corresponding controls matched for age and sex.
Definition of cases and controls
A case was defined as a child with airway complaints, who received a diagnosis of acute respiratory after visiting the health facility to seek treatment for ARI for the first time during that episode and who had not used antibiotics or antiviral medications in the previous 2 weeks. Control subjects were defined as children who had complaints other than respiratory complaints, had no complaints of an ARI in the prior 2 weeks, did not belong to the same household as the case, and had not used antibiotics or antivirals in the previous 2 weeks. Controls were recruited within the same facilities and matched for age and sex.
Sampling
The study enrolled a total of 261 participants who gave informed consent. These were parents/ guardians and their children. However, 5 cases did not find matching controls and therefore their data was not included in the analysis. A sub-group of 78 children were selected to obtain a nasopharyngeal and throat specimen for isolation and identification of viral and bacterial etiological agents.
Specimen collection
Throat swabs were collected from tonsillar areas using sterile cotton swabs on wooden applicator sticks for bacterial isolation. The throat swabs were placed immediately in Amies transport medium and sent the laboratory immediately at room temperature for bacterial culture. For viral agents, the nasopharyngeal swabs were collected by inserting the swab into one nostril straight back along the floor of the nasal passage for several centimeters until reaching the nasopharynx. The swabs were rotated gently for 5-10 seconds to loosen the epithelial cells and collect the sample. The nasopharyngeal swabs were inserted into viral transport medium and transported for further processing. The nasopharyngeal samples were stored at -80°C if not processed immediately until further analysis. These samples were subjected to respiratory virus detection using multiplex nested PCR. The laboratory investigations were done at the Kenya Medical Research Institute (KEMRI) laboratories in Nairobi.
Laboratory analysis and identification of aetiological agents
Bacteriological specimen (throat swab)
Specimen were primarily be inoculated on blood agar base supplemented with 5% sheep blood to isolate the bacteria such as Streptococcus pneumonia, Staphylococcus aureus, beta haemolytic streptococcus. Gonococci (GC) agar base with 2% haemoglobin and 1% isovitalex were used to allow isolation Haemophilus influenzae. MacConkey with 0.01% crystal violet was used for isolation of Gram-negative bacteria such as Klebsiella species associated with ARI. Biochemical tests were used for identification of the bacterial species following isolation.
Viral specimen (nasopharyngeal)
Nucleic acid extraction: Viral Ribonucleic acid (RNA) from the specimen was extracted by a commercial kit according to the manufacturer’s protocol (QIAamp MinElute Virus Spin Kit, Qiagen, Valencia, and CA). Briefly 140µl of sample was added to 560µl of viral lysis buffer, incubated at room temperature (15-25°C) for 10 minutes, then 560µl of molecular grade 100% ethanol was added and mixed by vortexing for 15 seconds. This was then centrifuged briefly to remove drops from inside the Eppendorf tube lid. From the lysed RNA, 630µl of RNA was then placed on to a spin column, spun at 6000xg, twice binding the RNA to the spin column. The RNA was washed twice, first with 500µl of wash buffer 1 (AW1) at 6000xg for 1 minute, then with 500µl of wash buffer 2 (AW2) at 20,000xg for 3minutes. The RNA was eluted from the spin column by adding 60µl elution buffer (AVE) and spinning at 6000xg for 1 minute to a 1.5ml Eppendorf tube. PCR was conducted using primer sequences outlined in Table 1 targeting different types of viruses.
Virus |
Primers |
Sequence (5’-3’) |
Gene |
Amplicon Size (bp) |
hRSV |
vrs 1 |
GGA ACA AGT TGT TGA GGT TTA TGA ATA TGC |
Nucleo capsid |
279 pb |
vrs P2 |
TTC TGC TGT CAA GTC TAG TAC ACT GTA GT |
|||
Influenza A Virus |
mia 1 |
CAG AGA CTT GAA GAT GTC TTT GCT GG |
Matrix protein |
212 |
mia 2 |
GCT CTG TCC ATG TTA TTT G |
|||
Influenza B Virus |
mib 1 |
AAA ATT ACA CTG TTG GTT CGG TG |
Matrix protein |
362 |
mib 2 |
AGC GTT CCT AGT TTT ACT TG |
|||
hMPV |
hmpv 1 |
CCC TTT GTT TCA GGC CAA |
Matrix protein |
416 |
hmpv 2 |
GCA GCT TCA ACA GTA GCT G |
|||
RT-PCR Parainfluenza Virus 1 |
PIS1+ |
CCG GTA ATT TCT CAT ACC TAT G |
Hemagglutinin- |
317 pb |
PIS1- |
CCT TGG AGC GGA GTT GTT AAG |
Neuraminidase |
||
Parainfluenza Virus 2 |
PIP2+ |
AAC AAT CTG CTG CAGCAT TT |
Hemagglutinin- |
507 |
PIP2- |
ATG TCA GAC AAT GGG CAA AT |
Neuraminidase |
||
Parainfluenza Virus 3 |
Para3.1 |
CTC GAG GTT GTC AGG ATA TAG |
Hemagglutinin- |
189 |
Para3.2 |
CTT TGG GAG TTG AAC ACA GTT |
Neuraminidase |
||
Internal Control |
GAPDH1 |
TCA TCC ATG ACA ACT TTG GTA TCG TG |
GAPDH |
564 |
GAPDH2 |
CTC TTC CTC TTG TGC TCT TG |
|||
Table 1 Primer sequences and target genes used
Reverse transcriptase real-time polymerase chain reaction: In order to detect and subtype influenza viruses from the samples, a real time RT-PCR protocol designed by World Health Organization (WHO) and the Centers for Disease Control (CDC) was used. Briefly, 5µl RNA was added to a 20µl master mix containing buffer, primers (both forward and reverse), probe, nuclease free water and an enzyme. This was carried on an AB 7500 Fast Real-Time PCR (Applied Biosystems). The cycling conditions for the real time RT-PCR protocol were: an initial cycle at 400C for 30 minutes for the reverse transcription of the RNA to develop the complimentary DNA (cDNA). This was followed by incubation at 94°C for 10 minutes to inactivate the reverse transcriptase and activate the Taq polymerase. This was followed by 40 cycles involving denaturation at 94°C for 15 seconds, and an annealing and strand extension stage at 55°C for 1minute.
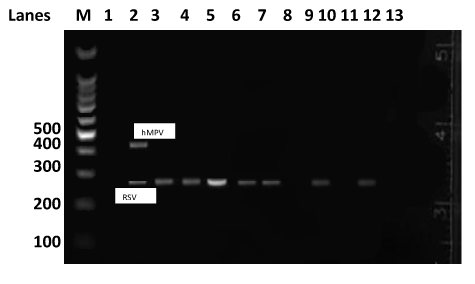
One-step reverse transcriptase polymerase chain reaction: The reverse transcriptase PCR (RT-PCR) procedure consisted of a single-step combining reverse transcription and PCR amplification, performed using the one-step RT-PCR kit from QIAGEN (Hilden, Germany). The reaction mixture contained 5µl of 5× RT-PCR buffer, 1µl of 0.4mM dNTPs, 1.25µl of each of the primers (forward and reverse primers in multiplex PCR for RSV/hMPV in one reaction, and a second reaction for Parainfluenza viruses (1 to 3) using a similar method as described by Bellau-Pujol et al.16 Briefly, the RT-PCR involved 13µl nuclease free water and 1µl of enzyme mix. A 2.5µl aliquot of the extracted sample RNA was added to give a final volume of 25µl. The cycling conditions for the RT-PCR were: an initial cycle at 50°C for 30 minutes for the reverse transcriptase to work followed by incubation at 94°C for 15 minutes to inactivate the reverse transcriptase and activate the Taq polymerase. This was followed by 40 cycles involving denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds and strand extension at 72°C for 1 minute. Finally, a final extention at 72°C for 10 minutes to completely fill in the amplification productswas carried out on AB 9700 PCR thermocycler (GeneAmp® Applied Biosystem). The PCR products were visualized under UV light after gel electrophoresis on 2% agarose gels and stained with ethidium bromide using an HP AlphaImager® (Alpha Immutech, South Africa; Figures 1 & 2).
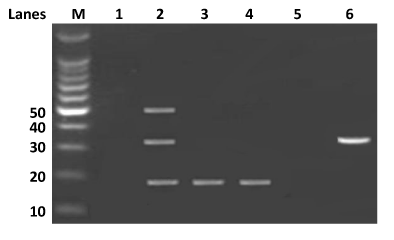
Figure 1 Detection of parainfluenza viruses multiplexes RT-PCR.
This assay was to differential parainfluenza types 1, 2 and 3 from the clinical specimen. Lane M=100bp DNA marker (Invitogen Cat74602-250). Lane 1 is an internal negative control, lane 2 has parainfluenza 1 (317bp), 2 (507bp), and 3 (189bp) positive controls. Lanes 3 to 6 represents field samples used in this study.
Ethical considerations
The study protocol was reviewed and approved KEMRI Scientific Steering Committee (SSC) and KEMRI Ethical Review Committee (ERC) prior to commencement of the study. Participants were asked to give written informed consent to participate in the study and participation was entirely voluntary. No invasive procedures were carried out and no adverse events were experienced during the study.
Statistical analysis
The data was analyzed using statistical package for social scientists (SPSS/PCTM) software. The data on etiology was analyzed using descriptive statistics and results expressed as rates and proportions. Chi square test and Odds ratio were used to test the associations of etiology and risk factors. P-value of less than 0.05 was considered significant.
Demographic characteristics
Caretakers who provided informed consent to participate in the study aged between 18 to 46 years with a median age of 27 years (Mean±SD; 27.6±5.6). The children were aged between 4 and 60 months with median age of 24 months (Mean±SD; 24.8±15.8). The children had a median weight of 3kgs (Mean±SD; 3±0.6). Most (59%) of the children enrolled in the study were males. No significant difference was found between cases and control’s age but a significant difference was found in children’s mean birth weights (p<0. 0001; Table 2).
Characteristics |
Total |
Cases |
Controls |
P |
||||||
Mean±SD |
Median |
Range |
Mean±SD |
Median |
Range |
Mean±SD |
Median |
Range |
||
Child Age (Months) |
24.8±15.8 |
24 |
4-60 |
24.5±16.1 |
23.5 |
4-60 |
25±15.4 |
24 |
4-60 |
0.8 |
Child Birth Weight (Kgs) |
3±0.6 |
3 |
2-7.8 |
3.1±0.6 |
3 |
2-7.8 |
3±0.6 |
3 |
2-7.5 |
<0.0001 |
Parent Age |
27.6±5.6 |
27 |
18-46 |
27.6±5.9 |
27 |
18-46 |
27.6±5.4 |
27 |
18-45 |
0.99 |
Table 2 Distribution of participants by age and weight
Viral and bacterial etiologies
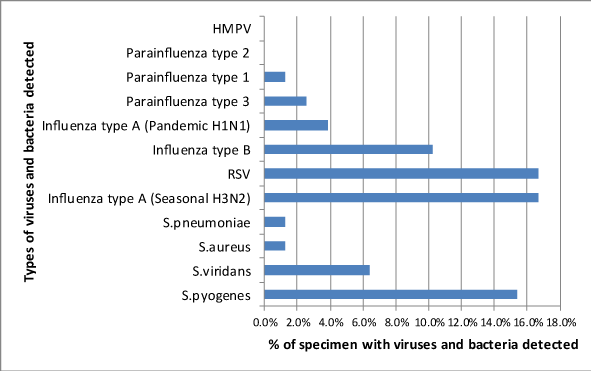
Bacteria were isolated from 19 of 78 (24.4%) throat swabs while at least one type of virus was detected in 35 out of 78 (44.9%) nasopharyngeal specimen (Figure 3). Among the bacteria isolated, Streptococcus pyogenes was most common, isolated in 13 out of the 78 (15.4%) throat swabs collected from the children followed by Streptococcus viridans, 5 (6. 4%). Other bacteria isolated in fewer specimens were Streptococcus pneumoniae and Staphylococcus aureus. Influenza type A virus was the most commonly isolated virus, detected in 21 out of 78 (26.9%) specimen followed by RSV, 13 (16.7%) and Influenza type B (10.3%) of the specimen. Among the Influenza type A viruses, seasonal Influenza A (H3N5) was more common, 13 (16.7%) than pandemic Influenza A (H1N1) which was detected in 3 (3.8%) of the entire clinical specimen. Other viruses detected in few numbers of clinical specimens were Parainfluenza type 1 and Parainfluenza type 3 while two and one clinical specimen respectively while Parainfluenza type 2 and hMPV viruses were not detected in any of the specimen (Figure 4). Representative photographic agarose electrophoresis display of multiplex PCR products parainfluenza viruses, RSV and hMPV are shown in Figures 1 & 2 below. Mixed agents were detected in 23 out of the 78 (29.5%) specimens. All the nineteen children who had bacterial agents isolated, had a viral co-infection. Bacteria and a single virus co-infection were the most predominant 19 (78.3%). One specimen each contained a mixture of bacteria and more than one virus while others contained a mixture of Influenza A and RSV, influenza B and RSV, Parainfluenza 1 and RSV and Parainfluenza type 3 and RSV (Table 3).
|
Co-infected Specimen |
|
Type of Agents |
Number |
Percentage |
Bacteria and single virus |
12 |
70.6% |
Bacteria and more than one virus |
1 |
5.9% |
Influenza type A and RSV |
1 |
5.9% |
Influenza type B and RSV |
1 |
5.9% |
Parainfluenza type 1 and RSV |
1 |
5.9% |
Parainfluenza type 3 and RSV |
1 |
5.9% |
Total number of co-infected specimen |
17 |
100% |
Although not statistically significant, cases were 2.5 more likely to have bacteria isolated than controls (OR=2.53; 95% CI, 0.81-7.92; p=0.17). All the children with bacteria isolated from their clinical specimen had viral pathogens detected as well (Table 4). Twenty five (32.1%) had viruses only among them were 16 (35.6%) of the cases and 9 (27.3%) of the controls. As shown in Table 4 below, viruses were more likely to be detected in cases than in controls, although the difference was not statistically significant (OR=1.5; 95% CI, 0.6-3.9; p=0.5).
Patient Type |
|||||
Agents Detected |
N=78 |
Cases n (%) |
Controls n (%) |
OR (95% CL) |
p |
Bacteria and viruses (mixed)* |
|||||
Yes |
19 (24.4) |
14 (22.2) |
5 (15.2) |
1 |
0.17 |
No |
59 (75.6) |
31 (68.8) |
28 (84.8) |
2.53 (0.81-7.92) |
|
Viruses Only |
|||||
Yes |
25 (32.1) |
16 (35.6) |
9 (27.3) |
1 |
0.47 |
No |
53 (67.9) |
29 (64.4) |
24 (72.7) |
1.5 (0.6-3.9) |
|
Viral Pathogens** |
|||||
Influenza Type A |
16 (40.0) |
15 (55.6) |
1 (7.6) |
0.063 (0.008-0.503) |
0.001 |
Influenza Type B |
8 (20.0) |
6 (22.2) |
2 (15.4) |
0.419 (0.079-2.224) |
0.296 |
RSV |
13 (32.5) |
6 (22.2) |
7 (53.8) |
1.75 (0.528-5.778) |
0.356 |
Table 4 Association of isolation rate of bacterial & viral agents among cases and controls
*All children with bacteria isolated had viruses as well.
**No HMPV and parainfluenza type 2 were detected while parainfluenza type 1 and 3 was isolated from one and two participants respectively and therefore was exempted from the analysis.
Table 5 shows the association age and sex with bacterial and viral agents’ detection from the cases and controls. Bacteria were isolated in higher proportion of females, 9 (32.1%) compared to males 10 (20%) respectively (OR=1.89; 95% CI, 0.585-6.165; p=0.231). On the contrary, viruses were detected in a higher proportion of male children, 24 (48%) than females, 12 (42.9%; p=0.81). Bacterial agents were isolated in a higher proportion of the children aged between 1-3 years, 11 (32.3%). This distribution was not significantly different (p=0.345). Viruses were detected in significantly higher proportion, 16 (61.5%) among the children aged up to one year of age than other higher age groups (p<0.03).
Bacteria Isolated |
||||
Category |
No (%) |
Yes (%) |
OR (95% CI) |
P |
Sex |
0.231 |
|||
Female |
19 (67.9) |
9 (32.1) |
1 |
|
Male |
40 (80.0) |
10 (20.0) |
1.89 (0.585-6.165) |
|
Child Age (Years) |
||||
0-1 |
21 (80.8) |
5 (19.2) |
1 |
0.345 |
>1-3 |
23 (67.7) |
11 (32.3) |
0.49 (0.14-0.67) |
|
>3 |
15 (83.3) |
3 (16.7) |
1.19 (0.25-5.8) |
|
Table 5 Association of age, sex and bacterial and viral agent’s detection from ARI cases and controls
Clinical characteristics
Children who presented with fever were two times more likely to have bacteria isolated than those without fever (OR=2.087; 95% CI, 0.381-11.417). Similarly those who had high respiratory rate were more likely to have bacteria isolated (OR=2.545; 95% CI, 0.44-14.585). Other clinical signs associated with isolation of bacteria were cough (OR=6.19; 95% CI, 0.708-54.157) and stridor (OR=5.0; 95% CI, 0.414-60.431). Children with high respiratory rate were 1.7 times more likely to have viruses detected than those with no reported high respiratory rate (OR=1.714; 95% CI, 0.28-10.479). Cough was also related with the detection of viruses among the cases (OR=2.827; 95% CI, 0.69-11.58). Fever as well as other clinical features did not show association with viral detection (Table 5).
Bacteria were isolated from 19 of 78 (24%) throat swabs of children under the study. Streptococcus pyogenes and Streptococcus viridans were the most predominant bacterial agents, isolated in 15.4% and 5(6.4%) of the throat swabs respectively. Streptococcus pneumoniae and Staphylococcus aureus were isolated in only a few specimens. Few studies done previously on etiology of ARI indicate the role of Streptococcus pyogenes (group A β-hemolytic streptococci) in causing Acute upper respiratory infection. Mahmud et al.,17 reported bacterial isolation rate of 31.4% among the study subjects with predominant bacterial isolates such as Staphylococcus aureus (12.4%) and Streptococcus pyogenes (9.8%). Mandell18 in another study reported Streptococcus pyogenes (11%) as the predominant bacteria in patients with ARI. A study in Netherlands by van Gageldonk-Laafeber et al.,2 isolated haemolytic streptococci in 41(8%) of the study subjects. Findings of this study agree with the previous study on Streptococcus pyogenes being the most important bacterial agent associated with ARI in children.
This study detected at least one ARI respiratory associated virus in 44.9% of the specimen from the children. The study findings compares to work of previous studies conducted in Kenya and elsewhere. A study conducted in Kilifi, Kenya, by Munywoki et al.,19 found that 66.6% children had at least one respiratory virus detected. Ahmed et al.,20 detected respiratory viruses in 49.8%of study subjects in a study conducted at a refugee camp in Kenya. Studies conducted elsewhere in the World revealed that respiratory viruses were common in children with ARI. Bharaj et al.,21 in a paediatric study in India reported a viral detection rate of 35.2%while Louie et al.,22 in study in San Francisco, California in the United States reported a respiratory detection rate of 38%. Another study conducted in Australia reported a viral detection rate of 69% in ARI suspected patient.23 A study conducted in children under five in Brazil reported the highest virus detection rate of 85.5%.24 The above studies report respiratory viruses’ detection rate of 35% to 86%, indicating the role of respiratory viruses as etiological agents of acute respiratory infections. The findings of the current study are consistent with findings found in other studies conducted within the country, in developing and in developed world especially the findings of Ahmed et al.,20 in Kenya who reported detecting viruses in 49.8% clinical specimen. However, it is important to note that not all possible respiratory viruses were tested. Compared to bacterial agents, viruses were more commonly isolated among the study subjects. These findings are in agreement with the conclusions made by van Gageldonk-Laafeber et al.,2 that reported that most ARI are viral.
Various respiratory viruses were detected in this study which included; Influenza A which was detected in majority (20. 5%) of the specimen, followed by RSV (16.7%) and Influenza B detected in 10.3% of the samples. Human Parainfluenza 1 and Parainfluenza 3 were detected in minority of the samples, 2.6% and 1.3% specimen respectively. Parainfluenza 2 and hMPV viruses were not detected in any of the patient samples. Gageldonk-Laafeber et al.,2 reported a predominant detection of Influenza and rhino viruses in a study in Netherlands. Findings of Ahmed et al.,20 detected predominantly RSV (12.5%) and others were hMPV in 5.7%; Parainfluenza in 9.4%; Influenza A, 9.7%; and Influenza B in 2.6%. Bezerra et al.,24 reported detection of RSV in 37% of the subjects and hMPV in 10% while Bharaj et al.,21 reported that out of 103 clinical specimen positive for respiratory viruses RSV contributed the majority (61, 59.2%). In other related studies, Kusel et al.,23 found RSV (10.9%) to be among the most common viruses while Munywoki et al.,19 reported RSV being detected in 24.4% of the study subjects.
Our study found a strong involvement of Influenza A, RSV and Influenza B among the study subjects. Among the influenza viruses, Influenza A accounted for 66.7% (16/ 24) compared to Influenza type B, 33.3% (8/ 24). These findings are in concurrence with the report of the WHO Global Influenza Surveillance and Response System (GISRS). According GISRS, during the epidemiological period January-September 2013, the period in during which this study was conducted (April to September 2013), among ILI cases reported, Influenza A accounted for 86% of all influenza viruses detected (728/ 845) and Influenza B represented 13% (117/ 845). Our study found that among the Influenza type A detected, seasonal Influenza type A (H3N2) was more predominant, 81.2% (13/16) compared to pandemic Influenza type A (H1N1). GISRS report indicated that Influenza A (H1N1) pdm09 transmission peaked around 9 weeks earlier than A (H3N2), resulting in a marked predominance of A(H1N1)pdm09 in the first 3 quarters of the season, with transition to A (H3N2) in the final quarter of the season.25 The findings of this study were consistent with the reports of the WHO GISRS in the southern hemisphere where more seasonal H3N2 was reported compared to pandemic H1N1 and Influenza B after the first three quarters of the season. Contrary to the findings of our study which detected Parainfluenza viruses in only 3.8% of the clinical samples and found no involvement of hMPV among the study participants, several previous studies reported Parainfluenza virus and hMPV in over 10% and over 5.0% subjects respectively and found little involvement Influenza A.19,21,24
The study reported mixed infection of virus and bacterial pathogens. A mixed infection was defined as the presence of a virus in combination another one or more other viruses or with one or more bacterial isolates. Mixed infections were reported in 21.8% of the patients with predominant bacterial and viral combination (70%). These findings are in agreement with previous studies which reported mixed infection of viral and bacterial pathogens associated with ARI.2 This indicates that a good proportion of patients presenting with ARI may be co-infected with viral and bacterial agents and hence management may require multi-agent screening to ensure proper management and successful resolution of ARI is achieved where multiple etiological agents are involved. Potential mechanisms of interaction of viral and bacterial pathogens have been reported in literature. Infection with a viral pathogen may predispose an individual to bacterial infection through a number of viral-bacterial infections;26,27 a few of which are explained below. McCullers28 explains one of the mechanisms reported in literature on viral-bacterial interaction on synergism between influenza virus and S. pneumonia. Although an influenza virus infection alone can be fatal, mortality increases dramatically when a bacterial super-infection occurs, as in the case of the ‘‘Spanish flu’’ pandemic in 1918-1919 when millions of people died, most from secondary Pneumococcal pneumonia. This is further underlined by animal experiments showing that death occurred in 35% and 15% of mice infected with either influenza virus or pneumococcus, respectively, where as 100% of mice infected with both pathogens simultaneously succumbed to infection within one day.29 Another mechanism of viral-bacterial interaction is viral predisposition and bacterial adherence in which explains that since attachment of a pathogen to mucosal surfaces is the first step towards respiratory disease, and viral infection alters the defense of the host epithelium in general, it has been postulated that viral presence may render the epithelium more susceptible to bacterial colonization.30 Respiratory viruses may also directly affect the immune system, for example by impairment of neutrophil function, decreased oxidative burst and enhanced neutrophil apoptosis, thereby increasing susceptibility to bacterial super-infection.31 Additionally, some strains of influenza virus infection may predispose to superinfection by S. aureus due to ineffective natural killer (NK) cell recruitment and activation.32 The findings would propose a future study on pathogenesis and viral-bacterial interaction may explain the mechanisms of interaction in our local population.
A breakdown of the most common pathogens in case patients and control subjects revealed that Influenza A viruses were the most common pathogens in case patients (55.6%; p=001). RSV was the most common pathogen among the control subjects (53.8%; Table 4). Findings of the study are in agreement with the study by Mandell18 reported that Influenza type A virus to be the most predominant virus in causing ARI (42%) while RSV was the most common viral agent in control (asymptomatic) subjects (17%). The detection of RSV in study subjects without symptoms is suggestive that asymptomatic subjects may harbor pathogens and could be a potential source of transmission of respiratory pathogens. Asymptomatic persons may act as unsuspected sources of infection.
Relating clinical presentation of the patients with viral and bacterial detection, children with fever (OR=2.087; 95% CI, 0.381-11.417), high respiratory (OR=2.545; 95% CI, 0.44-14.585), cough (OR=6.19; 95% CI, 0.708-54.157) and stridor (OR=5.0; 95% CI, 0.414-60.431) were more likely to have bacteria isolated while high respiratory rate (OR=1.714; 95% CI, 0.28-10.479) and cough (OR=2.827; 95% CI, 0.69-11.58) was more associated with viral isolation. Louie et al.,22 found association of fever with detection of viral agents (Influenza and RSV) while patients with RSV were more likely to have wheezing at examination than were others. In another study in Hong Kong, more children infected with Influenza A, adenovirus and mixed viruses had higher fever.14 This study did not find fever and wheezing to be significantly associated with viruses’ detection. The differences in the findings of this study could be attributed to the different study population and possibly involving physiological differences of the study subjects.
The study has shown the major viral and bacterial etiologies ARI in the study population was caused by influenza viruses and Streptococcus pyogenes respectively. This is information is important for the disease control program planning for surveillance and control of ARI in the population. Viruses were including RSV and Influenza were detected in control subjects without symptoms suggestive that asymptomatic subjects may harbor pathogens and could be a potential source of infection for acute respiratory infections.
We would like to thank all the staff at FITC health centre and Rongai health centres particularly Stephen Belio who assisted in the data collecting for this study. We would also like to thank the study subject from the two study site for their participation in the study. We also thank John Kiiru, Jane Muyodi and Symekher Samwel who assisted with the laboratory testing and also providing valuable technical input to this work.
Contribution of authors
Martin Matu (ECSA-HC and JKUAT), Gideon Kikuvi (JKUAT), Peter Wanzala (CPHR-KEMRI) and Mohamed Karama (CPHR-KEMRI) participated in idea conception and study design; Martin Matu and Samwel Symekher performed demographic and laboratory data collection; Martin Matu conducted data entry and analysis; Martin Matu Gideon Kikuvi, Peter Wanzala, Mohamed Karama and Samwel Symekher prepared the manuscript.
Authors declare that there is no conflict of interest.

©2014 Matu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.