Journal of
eISSN: 2373-437X


Mini Review Volume 5 Issue 4
Research Director, A&L Biologicals, Agroecological Research Services Centre, Canada
Correspondence: George Lazarovits, Research Director, A&L Biologicals, Agroecological Research Services Centre, London, Ontario, Canada, Tel 519-457-2575; 246
Received: May 19, 2017 | Published: July 26, 2017
Citation: Lazarovits G, Ali S, Kandasamy S, Saldias S (2017) A Road Map to Finding Microbiomes that Most Contribute to Plant and Soil Health. J Microbiol Exp 5(4): 00153. DOI: 10.15406/jmen.2017.05.00153
The interaction between plants and their associated microbes varies from being beneficial to neutral to deleterious. The development of ecological agriculture, where greatly less extraneous inputs will be used, requires us to identify and deploy microorganism that form beneficial relationships with crops and act to enhance their health and yields. Robust and inexpensive molecular techniques have allowed for rapid identification of key players and their interactions with plants but there is still a need to discover what factors regulate these interactions and to develop methods for delivering microbial products to the environments where they are needed. There is ample evidence that examining agroecosystems where production methods have resulted in exemplary high productivity of plants are the ideal conditions from which to isolate, identify and examine the factors that allow microorganisms to optimally exert their influences on host plants. With the introduction of aerial monitoring of crops, we could identify site specific locations within fields there were significant differences in the microbial interactions and that allowed us to examine factors associated with the variable productivity of plants within a field. Although we are still at the early stages of such studies there is a high probability that an agroecological approach will allow for the identification of the chemical, physical, environmental and microbiological factors that regulates plant-microbial interactions and to assess how such factors impact crop yields.
Keywords: pathogens, microorganisms, agriculture, chemical, physical, environmental
ISR, induced systemic resistance; TRFLP, terminal restriction fragment length polymorphism; DAPG, 2,4-diacetylphoroglucinol; NDVI, normalized difference vegetation index
Microbiology studies for the most part have focused on the impacts of microorganisms as pathogens or for their use in industrial production of valuable commodities. Today however, the primary focus is on identifying their role in ecosystem health and ecology. The very significant reduction in the cost and speed for molecular tools and sequencing continues to significantly increase our abilities to examine whole microbial communities and to identify their potential functions. In agriculture sector, results from such studies have considerably improved our knowledge to comprehend plant-microbe interactions and have provided revolutionary information as to the role these microbiomes play in plants’ health. The plant microbiome could potentially help its host by providing nutrients, producing phytohormones, synthesizing vitamins, detoxifying toxic compounds, stimulating plants’ induced systemic resistance (ISR), and protecting them from a variety of biotic and abiotic stresses, etc. The unravelling of the plant microbiome is changing agriculture practice and the concept of what is a “healthy plant”. The agromicrobial revolution primarily focuses on the optimal use of existing plants’ microbial companion in order to improve plant performance and agricultural ecosystem functioning. Agricultural production systems must be examined from an ecological approach, with crop productivity being related to ecosystem services. Plant-associated microorganisms are fundamental for plant health and productivity as they affect plant nutrition, metabolism, physiology and performance. While the negative impacts of microorganisms on agroecosystem performance remains important, a greater focus on their beneficial impacts deserves closer attention. However, it must be emphasized that such benefits are going to be realized slowly. Here we provide three examples of how plant-microbe interactions have been utilized over millions of hectares and why it took decades for their utility to be realized. Suggestions that we can change agroecosystems overnight will only lead to disappointments in the research results.
The most well understood and exploited trait in the plant-microbial interaction catalogue is nitrogen fixation by Rhizobium species. The ability of Rhizobia to make atmospheric nitrogen available to plants and significantly increase their yields has been known for over 150 years. It has been estimated that nitrogen fixation by legumes in natural ecosystems is in the range of 25-75 lb of nitrogen per acre per year whereas, in cropping systems it may be several hundred pounds per acre.1 Commercially available Rhizobium inocula have a nominal cost by comparison to their yield and environmental benefits. The nodules formed on legumes remains the only plant-microbial interaction that breeders recognize as being critical to the success of any new cultivar they aim to generate. While plant-microbe interactions of similar significance likely occur with many other crop species, the lack of any obvious phenotypic indicator (i.e. presence of nodules analogous to that of legumes representing plant-Rhizobium interactions) likely could have resulted in the loss of genetic traits in the plants required for a successful outcome of the interaction with a designated partner. Who knows how many of such genetic functions have been deleted through the millennia of breeding processes? Studies of the interactions between Rhizobia and their hosts have revealed that there is an exchange of numerous chemical signals and this is a form of molecular dialogue. Hundreds of genes are likely involved in regulating the interactions of the two main groups of molecules that are required for a successful interaction. These are the nod gene-inducing flavonoids from the plant and the lipochito-oligosaccharide Nod factors from the Rhizobia. We have no idea of the traits that regulate microbial associations in corn, wheat, or rice in old or modern cultivars. The finding that Rhizobia can also form endophytic associations with rice and are able to colonize all the internal tissues of plant suggests that such interactions can have a major role in plant fitness and productivity. Surprisingly, the interactions proved to be strain and variety specific2 indicating that the growth responses are indeed heritable traits. Large-scale field trials evaluating five rice varieties and seven Rhizobia strains over five seasons showed that bacterial treatments increased yield by up to 47% in farmers’ fields, with an average increase of 19.5%. This study exemplifies the critical importance for selection of appropriate isolates and specific crop cultivars for optimizing yield benefits. We need to appreciate the growers of the Nile Delta who had the wisdom to recognize that intercropping rice with legumes contributed to yield increases of their rice crop. However, it was the selection of this site for study by the researchers that allowed them to discover that the build up of the selected Rhizobia populations provided the critical benefit to the agroecosystem functions in this area.
Modern agriculture was designed to provide crop plants with the optimal levels of all extraneous necessities for growth and yield. This approach however, may also have compromised the microbial partners associated with the crop plants. In the development of sugarcane as a bioethanol stock in Brazil, cultivars with high yield potentials were selected based their ability to perform at sites where fertilizers were never used.3 The best yielding cultivars were subsequently found to be colonized by numerous species of endophytic bacteria that provided the plants with nutrients and growth factors required for high yields, but without the need for large quantities of extraneous fertilizers. Sugarcane production in Brazil uses 50kgN/ha vs. the 350kg of N/ha used in the USA.3 It is not known if this approach can be used to select other crops plants to provide acceptable yields in the absence of large inputs of fertilizers, but the concept should be evaluated with other crops. The development of the Brazilian sugarcane industry also suggests that the most likely areas to find beneficial microorganisms within a field are at sites with exceptionally high yields. Our efforts thus have focused on examining sites where production practices have created exceptional yields as means to identify if, and what beneficial role microbiomes play in such instances.
Everything we do to grow plants, including the type of plants, impacts soil health and its microbiology. The development of disease-suppressive soils for take-all disease of wheat, pioneered by James Cook,4 is an example of how soil and plant health can be built through microbiology. Cook’s research showed that continuous cropping of wheat for five or more years reduced or eliminated take-all disease of wheat. When a suppressive soil was added at low rates to a soil where disease was present disease development was curtailed, even in the presence of the pathogen. Suppressiveness was shown to be caused by a build-up of fluorescent pseudomonad bacteria capable of synthesizing a diversity of fungicidal compounds (up to five antibiotics or antibiotic-like substances including 2,4-diacetylphoroglucinol, DAPG) that inhibit the pathogenic fungi.4 Crop rotations that reduced populations of this specific group of fluorescent pseudomonads eliminated suppressiveness, whereas those that increased them enhanced crop health. The focus for both soil and wheat health is now to design rotations where bacteria populations are increased or at least maintained at the suppressive level. Similar disease suppressive systems have been found for other disease and crops and most are biologically based. We identified a grower in Southern Ontario (Mr. Glenney, Haldimand, ON, G-site) who developed a cropping system where strips of corn and soybeans are planted in the exact same place on alternating years using a no-till production system and precision planters. While no obvious benefits in yield were obtained for the first five years, by the sixth-year his yields increased and after twenty years plateaued at about 300bu/A in a region where the average yields are 150 bu/A. The grower wondered if he created a disease suppressive soil for corn. We used this site as a model farm to identify if the yield response maybe associated with biological factors derived from the cultural practices being utilized. We planted the same corn variety at a conventional farm (H site) and at the G site over two years and tracked changes in plant and soil chemistry and biology.5 Bacterial and fungal communities of the soil from roots (rhizosphere), washed roots, and sap from the stem were studied at three-successional plant developmental stages, (30,60 and 90 days after planting) using terminal restriction fragment length polymorphism (TRFLP) analysis, which is a fast, reproducible, inexpensive, and, robust molecular fingerprinting technique (Figure 1) that has been used to determine the microbial diversity in a variety of environmental samples.6–11 TRFLP based molecular fingerprinting can not only distinguish the microbial communities, but it can predict their composition based on the fragment sizes obtained. Our initial objectives were to determine when, where, and how to look using the most rapid and inexpensive technique (i.e. TRFLP). More thorough molecular techniques could then be carried out on the most relevant samples. TRFLP analysis revealed significant temporal restructuring and dynamic alterations in the bacterial communities in all parts of the corn plant but the greatest differences between G and H sites bacterial populations occurred in the stem sap by day 60 (Figure 2) and persisted until day 90. The plant sap microbiome became our test of choice for assessing microbial diversity in subsequent large scale field trials. Fungal diversity differed significantly only in rhizosphere soils of the two sites and this was present at all sampling times. It appears that bacterial endosphere communities are most affected by the crops’ physiological development, while the fungi are apparently most impacted in the rhizosphere by tillage practices.
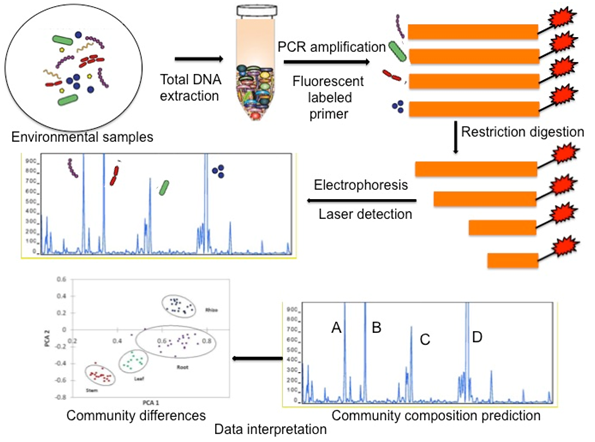
Figure 1 Illustration of molecular fingerprinting using terminal restriction fragment length polymorphism (TRFLP) analysis.

Figure 2 Bacterial communities of corn rhizosphere soil, root and sap samples at 60 Days after planting from high and low productive sites. Principal Component Analysis plots were constructed using the Terminal Restriction Fragment Length Polymorphism profiles (Islam et al.5 Unpublished data).
With a level of confidence that the microbiomes in corn sap can be readily differentiated from high and low yielding sites by 60 days after planting we went on to test plants harvested randomly across several corn fields. The results were disappointing in that we found no statistically significant differences in any of the samples for any field. By chance we received corn samples from fields that were sampled from specific sites within a field based on normalized difference vegetation index (NDVI) images collected by a drone. Here we found highly significant differences in the sap microbiomes from plants associated with stressed sites versus sites with highly vigorous plants (Figure 3A). We now have results collected from over 40 fields using NDVI images to sample high and low production sites. In most cases, significant differences were identified in their bacterial communities using TRFLP analysis. NDVI images indicate that almost all fields have poor, mediocre and high producing zones, where yields can vary from 75 to 350 bu/A. These zones might each account for about 1/3 of the farm area. The bacterial microbiomes within a specific zone in a field overall reflected yield expectations but why they were so differentially distributed or what they were doing within the plants remains to be determined. The results however, explain our earlier finding that when you sample across a field you can expect to identify the average corn microbiome. However, if you can partition the field to productive and underperforming plants you will find significant differences in communities that occupy various niches of the plants. Such variances can be seen in even very small plots (Figure 3B). The good news for growers is that one third of their fields are already producing well. We are currently analyzing the interaction between various plant and or soil physical, chemical, and biological factors related to plant and soil health and the productivity. We have not done justice to fungal populations in our studies, but do realize that they may have even greater impacts of crop productivity than the bacteria. The most obvious set of samples are being analyzed by high-throughput sequencing to examine the exact nature of the populations. Ultimately, our interest is to identify the physiological functions associated with the microbiomes that support plant vigour and yield. The major lesson here however, is that when you sample across a large population of plants you get the microbiomes of the average. We have established robust culture collection of microbes related to higher productive sites. It is very foreseeable that in the future we will have unique biofertilizers that will be formulated for broad spectrum functions for crop productivity. We already know that microbes can supply plants with nutrients of all types, protect them from diseases, and environmental stresses. How to introduce these into the plant ecosystem will be a major challenge, but first we need to know the conditions they require to thrive. Once we know this, we will be able to better manage agricultural practices that shift gene populations from the negative to the positive potentials.
We thank the Grain Farmers of Ontario for financial support of aspects work in this report. Funding for several projects mentioned here has also been provided by Agriculture and Agri-Food Canada through the Canadian Agricultural Adaptation Program (CAAP), the Growing Forward II program, The Ontario Genomics Institute and NRC-IRAP.
The authors declare that there is no conflict of interest.

©2017 Lazarovits, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.