Journal of
eISSN: 2373-437X


Research Article Volume 7 Issue 1
1Institute of Microbiology, Beijing Forestry University, China
2School of Economics and Management, Beijing Forestry University, China
3College of Life Science and Technology, Yangtze Normal University, China
Correspondence: Xiao-Hong Ji, Institute of Microbiology, Beijing Forestry University, PO Box 61, Beijing 100083, China, Tel 86-10-62336309
Received: December 30, 2018 | Published: January 18, 2019
Citation: Chen Q, Wang L, Du P, et al. A new species of Wrightoporiopsis (Russulales, Basidiomycota) and a key to accepted species in the genus. J Microbiol Exp. 2019;7(1):29-33. DOI: 10.15406/jmen.2019.07.00236
A new species of Wrightoporiopsis, W. irregularis sp. nov, is described and illustrated from southern China. It is characterized by annual, pileate, imbricated and sulphur yellow basidiocarps, irregular hymenophore varying from poroid to hydnoid, a monomitic hyphal structure in context but dimitic in the trama, generative hyphae bearing clam connections, indextrinoid skeletal hyphae, the absence of gloeocystidia, cystidia and gloeoplerous hyphae, the presence of fusoid cystidioles, ellipsoid, thin-walled, finely asperulate, strongly amyloid, and acyanophilous basidiospores measuring 2.8–3.3×2.2–2.5μm. Phylogenetic analysis based on the combined ITS (internal transcribed spacer region) and nLSU (the large nuclear ribosomal RNA subunit) dataset demonstrated W. irregularis is a new lineage in Wrightoporiopsis.
Keywords: hericiaceae, taxonomy, wood-inhabiting fungi
Wrightoporiopsis YC Dai, Jia J Chen & BK Cui, typified by W. neotropica (Ryvarden) YC Dai, Jia J Chen & BK Cui, was recently established by Chen et al.1 Some of taxa in the genus were previously treated under Wrightoporia Pouzar.2–4 However, Phylogenetic analysis demonstrated that Wrightoporiopsis is distant from Wrightoporia sensu stricto, and these two genera in fact belong to two families, Hericiaceae and Wrightoporiaceae, respectively.1 Wrightoporiopsis is characterized by pileate, yellow to yellowish-brown basidiocarps, a dimitic hyphal system with generative hyphae bearing clamp connections, skeletal hyphae usually dextrinoid, basidiospores ellipsoid to subglobose, hyaline, finely asperulate, strongly amyloid, and causing a white rot.1
During a field trip in Hainan Province of southern China, a yellowish specimen with poroid to hydnoid hymenophore was collected, it has a dimitic hyphal structure with generative hyphae bearing clamp connections, and asperulate, amyloid basidiospores, so it belongs to Wrightoporia sensu lato based on these morphological characters, and was not recorded in China.5 After phylogenetic analysis of ITS+nLSU sequences and re-examination morphology in laboratory, it turn out to represent a new species of Wrightoporiopsis. In this paper its illustrated description is given and an identification key to accepted species of Wrightoporiopsis is provided.
Morphology
The studied specimens are deposited in the herbaria of the Institute of Microbiology, Beijing Forestry University (BJFC). Morphological descriptions are based on field notes and herbarium specimens. Microscopic analyses follow Chen et al.,1 and Dai.6 Special color terms follow Anonymous7 and Petersen.8 In the text, the following abbreviations were used: KOH stands for 5% potassium hydroxide, CB stands for Cotton Blue, CB– stands for acyanophilous, IKI stands for Melzer’s reagent, IKI– stands for negative in Melzer’s reagent, IKI+ stands for amyloid in Melzer’s reagent, L stands for arithmetic average of all spore length, W stands for arithmetic average of all spore width, Q for L/W ratio, n (a/b) stands for measured from given number of spores (a) number of specimens (b).
Molecular phylogeny
The genomic DNA were obtained from dried specimens using the CTAB rapid plant genome extraction kit (Aidlab Biotechnologies, Co., Ltd., Beijing) following the manufacturer's instructions.1,9 The internal transcribed spacer (ITS) regions were amplified with the primers ITS4 and ITS5,10 and the nuclear large subunit (nLSU) ribosomal RNA gene regions with the primers LR0R and LR7.11 The PCR procedure for ITS and nLSU was follows Chen.1 The amplicon purified and sequenced by the Beijing Genomics Institute, China with the same primers as in amplifications. All newly generated sequence was deposited in GenBank (http://www.ncbi.nlm.nih.gov).
In addition to the newly generated sequences, additional ITS and nLSU sequences of Wrightoporiopsis and related species from previous studies1 were obtained from GenBank (Table 1) to explore the phylogenetic position of our specimen. All sequences were aligned using ClustalX v.1.8312 and manually adjusted in BioEdit.13 Before the phylogenetic analysis, ambiguous regions at the beginning and the end of the alignment were deleted and gaps were manually adjusted to optimize the alignment. The edited alignment was deposited at TreeBase (http://purl.org/phylo/treebase; submission ID 23041).
Phylogenetic analysis was following to previous studies.1,14 Maximum parsimony (MP), Bayesian inference (BI) and Maximum likelihood (ML) methods were used to perform the phylogenetic analysis. The three phylogenetic methods resulted in similar topologies for each dataset. Thus, only the topology from the MP analysis is presented. Branches that received bootstrap support from maximum parsimony (MP), maximum likelihood (BS) and Bayesian posterior probabilities (BPP) greater than or equal to 85% (MP and BS) and 0.95 (BPP) were considered as significantly supported.
Phylogenetic analyses
A total of 60 ITS (30) and nLSU (30) sequences included sequences from 31 fungal collections representing 16 species (Table 1) in this study, were used in the phylogenetic analyses. The alignment, generated by the ITS+nLSU dataset, contained 2147 characters. MP tree yielded four similar topologies (TL=1496, CI=0.606, RI=0.816, RC=0.494, HI=0.394). BI resulted in a similar consensus tree as the MP tree achieving an average standard deviation of split frequencies <0.01 after 2.5 million generations. ML tree also resulted in a topology similar to that with MP tree, and so only show the MP tree. BT values (≥80%) and BPPs (≥0.95) are shown at the nodes (Figure 1).
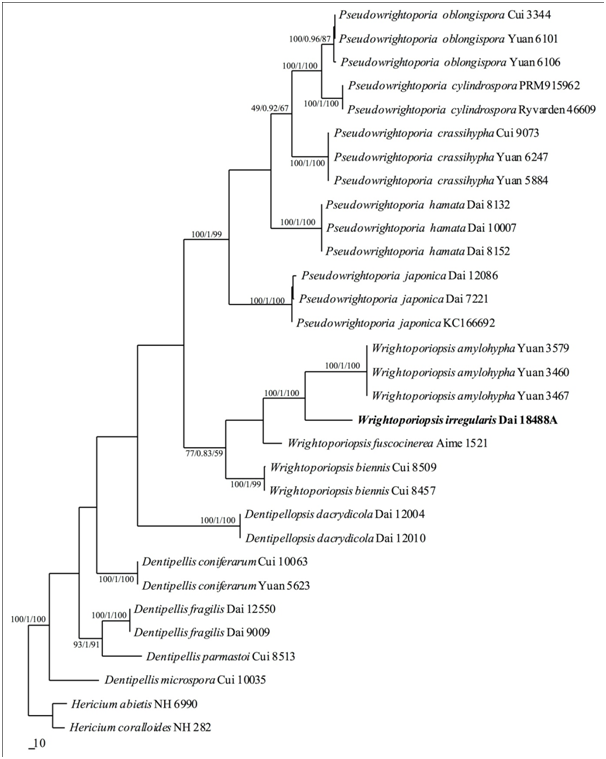
Figure 1 Strict consensus tree illustrating the phylogenetic position of Wrightoporiopsis irregularis, generated by maximum parsimony method based on ITS+nLSU sequence data. The topology is from the maximum parsimony analysis along with statistical values from the maximum parsimony, Bayesian inference analyses and maximum likelihood (bootstrap values and Bayesian posterior probabilities simultaneously not less than 80% and 0.9, respectively) at the nodes.
|
Taxa |
Sample no. |
Locality |
GenBank accession no. |
|
|
ITS |
nLSU |
|||
|
Dentipellis coniferarum |
Cui 10063 |
China |
JQ349106 |
JQ349092 |
|
Yuan 5623 |
China |
JQ349107 |
JQ349093 |
|
|
D. fragilis |
Dai 12550 |
China |
JQ349110 |
JQ349096 |
|
Dai 9009 |
China |
JQ349108 |
JQ349094 |
|
|
D. microspora |
Cui 10035 |
China |
JQ349112 |
JQ349098 |
|
D. parmastoi |
Cui 8513 |
China |
JQ349113 |
JQ349099 |
|
Dentipellopsis dacrydicola |
Dai 12004 |
China |
JQ349104 |
JQ349089 |
|
Dai 12010 |
Franc |
– |
JQ349090 |
|
|
Hericium abietis |
NH 6990 |
Canada |
AF506453 |
AF506453 |
|
H. coralloides |
NH 282 |
Sweden |
AF506459 |
AF506459 |
|
Pseudowrightoporia crassihypha |
Cui 9073 |
China |
KM107871 |
KM107890 |
|
Yuan 5884 |
China |
KM107872 |
KM107891 |
|
|
Yuan 6247 (holotype) |
China |
KM107873 |
KM107892 |
|
|
P. cylindrospora |
0810/1a |
USA |
GU594161 |
KJ807078 |
|
Ryvarden 46609 |
USA |
KJ513290 |
KJ807079 |
|
|
P. hamata |
Dai 8132 |
China |
KM107868 |
KM107887 |
|
Dai 8152 (holotype) |
China |
KM107869 |
KM107888 |
|
|
Dai 10007 |
China |
KM107870 |
KM107889 |
|
|
P. japonica |
Dai 7221 |
China |
FJ644289 |
KM107882 |
|
Dai 12086 |
China |
KJ513293 |
KM107883 |
|
|
KUC 20110908 |
Korea |
KC166692 |
KC166692 |
|
|
P. oblongispora |
Cui 3344 |
China |
KM107865 |
KM107884 |
|
Yuan 6101 (holotype) |
China |
KM107866 |
KM107885 |
|
|
Yuan 6106 |
China |
KM107867 |
KM107886 |
|
|
Wrightoporiopsis amylohypha |
Yuan 3460 |
China |
KM107875 |
KM107894 |
|
Yuan 3467 |
China |
KM107876 |
KM107895 |
|
|
Yuan 3579 (holotype) |
China |
KM107877 |
KM107896 |
|
|
W. biennis |
Cui 8457 |
China |
KJ807066 |
KJ807074 |
|
Cui 8506 (holotype) |
China |
KJ807067 |
KJ807075 |
|
|
W. fuscocinerea |
Aime 1521 (holotype) |
Guyana |
KM107897 |
– |
|
W. irregularis |
Dai 18488A (holotype) |
China |
MH626487 |
MH626488 |
Table 1 Specimens and GenBank accession number of sequences used in this study
New sequences produced by this work are in bold.
Wrightoporiopsis irregularis YC Dai, Q Chen & XH Ji, sp. nov. (Figure 2 & Figure 3)
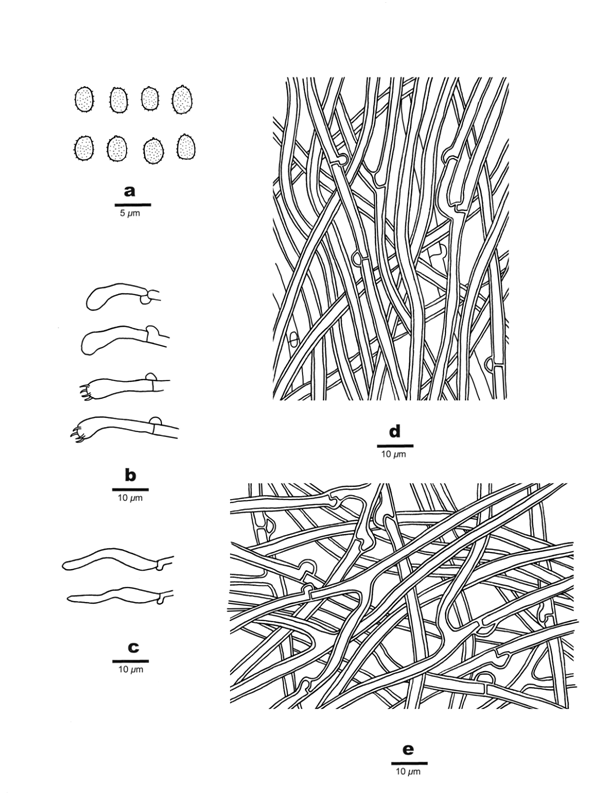
Figure 3 Microscopic structures of Wrightoporiopsis irregularis (Holotype). A: Basidiospores. B: Basidia and Basidioles. C: Cystidioles. D: Hyphae from trama. E: Hyphae from context.
Diagnosis: Differs from other Wrightoporiopsis species by its sulphur yellow to luteous basidiocarp, poroid to hydnoid hymenophore, a monomitic contextual hyphal structure, indextrinoid skeletal hyphae in trama, the absence of gloeocystidia and gloeoplerous hyphae.
Type: China, Hainan Province, Ledong County, Jianfengling Nature Reserve, on dead angiosperm tree, 26 April 2018, Dai 18488A (BJFC, holotype).
Etymology: Irregularis (Lat.): referring to the species has irregular hymenophore.
Description: Basidiocarp annual, pileate, imbricated, separable from substrate, soft and without odour or taste when fresh, corky when dry. Pilei more or less conchate, laterally fused, projecting up to up to 1cm, 3cm wide and 8mm thick at base. Pileal surface sulphur yellow when fresh, becoming pale luteous when dry, velutinate, azonate; margin blunt. Hymenophores sulphur yellow to buff when fresh, saffron to luteous when dry; margin distinct, concolorous with pileal surface, up to 1mm wide; hymenophore very irregular, poroid to sinuous when juvenile, becoming distinct hydnoid, pores or spines 2~4 per mm. Context concolorous with pileal surface, corky, up to 5mm thick. Tubes or spines concolorous with hymenophore, corky, up to 3mm long.
Hyphal system monomitic in the context, dimitic in the trama; generative hyphae bearing clamp connections; all hyphae IKI–, CB–, frequently encrusted by yellowish crystals; tissues becoming bloody red in KOH. Generative hyphae in context hyaline, thin- to thick- walled with a wide lumen, moderately branched, frequent bearing clamp connections, loosely interwoven, 2.5~4μm in diam. Generative hyphae in tubes hyaline, thin- to thick-walled, moderately branched and frequently bearing clamp connections 2.5~4μm in diam; skeletal hyphae frequent, hyaline, thick-walled with a wide lumen, rarely branched, flexuous, interwoven, 3~4.5μm in diam. Fusoid cystidioles present, hyaline, thin-walled, 22~28×4~6μm; basidia clavate, bearing four sterigmata and a basal clamp connection, 22~27×4~5μm; basidioles in shape similar to basidia, but slightly smaller. Basidiospores ellipsoid, hyaline, thin-walled, finely asperulate, strongly IKI+, CB–, (2.6~)2.8~3.3(~3.5)×(2~)2.2~2.5(~2.9)μm, L=3.09μm, W=2.33μm, Q=1.34 (n=30/1).
Previously five species of Wrightoporiopsis were reported: W. amylohypha YC Dai, Jia J Chen & BK Cui, W. biennis (Jia J Chen & BK Cui) YC Dai, Jia J Chen & BK Cui, W. fuscocinerea YC Dai, Jia J Chen & BK Cui, W. neotropica (Ryvarden) YC Dai, Jia J Chen & BK Cui, and W. roseocontexta (Ryvarden & Iturr.) YC Dai, Jia J Chen & BK Cui.1 W. amylohypha and W. biennis resembles W. irregularis by more or less yellowish hymenophore, tissue become red or brown in KOH, and distribution in China. However, W. amylohypha has regular poroid hymenophore with pores 5–6/mm, dextrinoid tramal skeletals and the presence of gloeoplerous hyphae.1 W. biennis has a biennial growth habit, regular poroid hymenophore with pores 6–9/mm, dextrinoid skeletal hyphae, the presence of gloeoplerous hyphae, and subglobose to broadly ellipsoid, cyanophilous basidiospores measuring 3.3~4×2.6~3.5μm.2 W. fuscocinerea can be distinguished from W. irregularis by its perennial and resupinate basidiocarp, regular poroid hymenophore with pores 8–10/mm, dextrinoid skeletal hyphae, the presence of gloeocystidia, and subglobose basidiospores measuring 3~4×2.5~3.5µm.2 W. neotropica is different from W. irregularis by resupinate basidiocarps, regular poroid hymenophore with pores 6–8/mm, dextrinoid skeletal hyphae, the absence of cystidioles and distribution in tropical America.3 W. roseocontexta is similar to W. irregularis by sharing indextrinoid skeletal hyphae, but differs in having resupinate basidiocarps, regular poroid hymenophore with pores 8~10/mm, the absence of cystidioles, globose basidiospores measuring 3~4µm in diam, and distribution in tropical America.4
Key to species of Wrightoporiopsis
1 Skeletal hyphae non-dextrinoid······································································································· 2
Skeletal hyphae dextrinoid··············································································································· 3
2 Hymenophores poroid, olivaceous brown, pores 8–10/mm; basidiospores globose, 3–4μm in diam W. roseocontexta
Hymenophores poroid to hydnoid, sulphur yellow to luteous, pores or spines 2–4/mm; basidiospores ellipsoid, 2.8–3.3×2.2–2.5μm W. irregularis
3 Basidiocarps pileate; contextual hyphae amyloid···························································· W. amylohypha
Basidiocarps resupinate to effused-reflexed; contextual hyphae inamyloid·············································· 4
4 Basidiocarps annual; basidiospores ellipsoid···································································· W. neotropica
Basidiocarps biennial to perennial; basidiospores subglobose to globose··················································· 5
5 Gloeocystidia present, gloeoplerous hyphae absent························································· W. fuscocinerea
Gloeocystidia absent, gloeoplerous hyphae present·································································· W. biennis
We would like to express our deep thanks to Prof. Yu-Cheng Dai (Beijing Forestry University) who allowed us to study his specimens. The research is supported by the Natural Science Foundation of China (Project No. 31750001).
Authors declare that there is no conflict of interest.

©2019 Chen, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.