Journal of
eISSN: 2373-437X


Borrelia organisms were harvested from blood culture and tested using single primers and loop mediated isothermal PCR (LAMP). Samples were harvested at either 6 days or at 8 weeks and DNA was extracted using one of three methods. In addition, pre-processing steps included Liberase treatments to release organisms from any residual attached collagen and human DNA removal. Combinations of processes were used to determine the best procedures for harvesting organisms, processing the pellet, and extracting DNA prior to using PCR. Of the total number of positive signals obtained, 9 of 45 (20%) gave positive signals when using plasmid primers specific for B. burgdorferi, 14 of 40 (35%) were positive when using OspC primers, and 25 of 47 (53%) were positive when using LAMP primers for the DNA that codes for 16S rRNA.
Keywords: LAMP, Borrelia, liberase
LAMP, loop mediated isothermal PCR; ALSI, advanced laboratory services inc.; CLSI, clinical laboratory standards institutes
The causative agent of Lyme disease, Borrelia burgdorferi, has proven to be difficult to culture in the laboratory. While organisms from ticks are routinely cultured in research labs using BSK media, the human pathogen found in the bloodstream is difficult to culture in artificial media. Advanced Laboratory Services, Inc. (ALSI) uses a modified blood culture media containing BSK-H, 12% rabbit serum and a collagen matrix.1 The culture media is inoculated with the patient’s blood and both short term (6 days) and long term cultures (8 to 16 weeks) are established. The 8 week to 16 week culture contains the collagen matrix. The use of this collagen matrix enhances the growth of Borrelia in the artificial laboratory culture media.
The positive control that is used to monitor these cultures is B. burgdorferi B31 ATCC 35210, originally isolated from an Ixodes dammini tick in New York. It is the type strain for B. burgdorferi, does not contain lipopolysaccharide, survives in experimentally infected human blood, is known to cause Lyme disease, is cultured at 34°C in BSK-H media under microaerophilic conditions, grows at three to five days, and its genome has been sequenced. The negative control, Escherichia coli ATCC 25922, is designated FDA strain Seattle 1946 Serotype 06, Biotype 1. It does not produce verotoxin and is used as a Clinical Laboratory Standards Institute (CLSI) strain for antimicrobial testing and for microbial identification in automated testing devices.
Investigators have used PCR to target Borrelia species found in ticks. These methods have included the use of nested primers, single tube nested primers,2 non-nested primers, and real time PCR to amplify Borrelia DNA. Such techniques were used to amplify DNA from ticks found in Norway, Germany and Austria. The genetic targets in these studies included intergenic spacer regions, OspA and the flagellin gene.2-4 LAMP technology has been used to target organisms directly from blood and to identify organisms grown in the laboratory.5,6
In our study, LAMP was used to amplify small amounts of DNA in the presence of inhibitors from patient blood cultures. LAMP primers were designed to detect DNA that codes for the 16S ribosomal unit of B. burgdorferi. Two other primer constructions were also tested on prepared samples. These primers were directed against the DNA that codes for the surface antigen OspC7 and DNA specific for B. burgdorferi that is carried on a stable plasmid (personal communication, Radhey Gupta). Samples were harvested from culture as described by Sapi et al.1
Treatment of pellets using Liberase TL
Liberase TL was used to release the organisms from any collagen that might be attached to the organism resulting from the harvesting process. Liberase TL was prepared using a stock solution of 2.5mg/mL, the equivalent of 13U/mL. 6µL of stock solution were added to 5mL PBS. This resulted in a final concentration of 2 U/mL in 1 X PBS which was added to the pellet in the tube and incubated for 4 min at 37°C. Following the incubation period, 25µL of Proteinase K was added to the tube to stop the reaction. Tubes were stored at -20°C until further use.
Removal of human DNA and RNA
Prior to extracting Borrelia DNA from the patient samples, human DNA was removed.8 This method used Oxgall (5%) to rupture mammalian cells and micrococcal nuclease to inactivate mammalian DNA. Controls were performed to ensure that Borrelia DNA would remain intact during this process. Concentrations of Oxgall at 1%, 1.5%, 2%, 5%, and 10% in PBS were mixed with B. burgdorferi N40 at a concentration of 2.6 x 107 cells, incubated at 37°C for 10 minutes, and densities were then read in a counting chamber.
To remove human DNA from patient samples harvested from blood culture,8 200µL of the Liberase treated sample was mixed with 200µL 10% Oxgall (prepared in Type 1 autoclaved water) for a final concentration of Oxgall at 5%. The tubes were incubated at room temperature for 10 minutes, and then centrifuged at 13,000 rpm for 5 minutes and the supernatant discarded. The pellet was resuspended in100µL of 0.1µM CaCl2 and 5µL (103 gel units) micrococcal nuclease and incubated at 37°C for 10 minutes. Following centrifugation at 13,000 rpm for 5 minutes, the pellet was resuspended in 200µL 1 X PBS and 10µL molecular grade Proteinase K was added. Incubation for 5 minutes at room temperature was performed to inactivate residual micrococcal nuclease before extracting the DNA.
Extraction of bacterial DNA using the MagJet Kit
DNA was extracted from samples using the MagJet Genomic DNA Kit (Cat. # K2721 Thermo Fisher Scientific, Life Technologies). This kit is designed for DNA extraction from bacteria, blood and tissues among other biological materials. Instructions were followed per the manufacturer.
Extraction of DNA using the Qiagen Kit
Bacterial DNA was also extracted using Cat # 51306 manufactured by Qiagen. This kit is designed to harvest both chromosomal and plasmid DNA from bacteria. The instructions supplied by the manufacturer were followed for this process.
Extraction of DNA using the MP Biomedical Fast Prep 24 semi-automated extractor
Tubes and processes for isolation of DNA from bacteria in soil samples were used as suggested by the manufacturer.
Harvesting time intervals
As designated in Table 3, harvesting from positive cultures was performed at various points in the life cycle of the culture. Harvest time points were 6 day positive and harvested at 6 days, 6 day positive and harvested at 8 weeks, eight week positive and harvested at 8 weeks.
PCR using OspC primers
Primers directed against the OspC gene were developed by Qiu et al.7 From this publication, primer designations OC6 F+ and OC602 R- were used. The master mix was prepared using 1-25ng of template, 20µM of each dNTP, PCR buffer, 1.5mM MgCl2 400 pM of each primer, 1.5 U of Taq, and 5% glycerol. The cycling conditions were 95°C for 5min, 95°C for 30sec, 50.5°C for 30sec, 72°C for 1min 30sec, repeat from step 2 for 39 times, and 72°C for 5min and 4°C keep. Two complete PCR cycles were run using the same primers. The size of the amplicon was 600 base pairs and amplicon size was confirmed by running a 2% agarose gel and using the 100 to 1000 bp ladder. Control DNA from B. burgdorferi B31 ATCC 35210 and E. coli ATCC 25922 were used with each run to validate the master mix conditions and cycling conditions.
PCR using B. burgdorferi plasmid gene primers
Primers directed against stable plasmids carried by B. burgdorferi were obtained from Dr Radhey S. Gupta, McMasters University, Hamilton, Ontario, Canada. These primers targeted the organism at the genus level. The forward primer was 5’-CTTGTCAAACTTATCAAATAGC-3’ and the reverse primer, 5’-AAATATGAATCATAAAAATCG-3’. Master mix was prepared using 1-25 ng of template, 20µM of each dNTP, PCR buffer, 1.5mM MgCl2 400 pM of each primer, 1.5 U of Taq, and 5% glycerol. The cycling conditions were 95°C for 5min, 95°C for 30sec, 55°C for 30sec, 72°C for 1min 30sec, repeat from step 2 for 39 times, and 72°C for 5 min and 4 °C keep. Two complete PCR cycles were run using the same primers. The size of the amplicon was 296 base pairs and amplicon size was confirmed by running a 2% agarose gel and using the 100 to 1000 bp ladder. Control DNA from B. burgdorferi B31 ATCC 35210 and E. coli ATCC 25922 were used with each run to validate the master mix conditions and cycling conditions.
PCR using LAMP 16S rRNA primers for B. burgdorferi
Ten LAMP primers were designed to target the DNA that codes for the16S rRNA region of Borrelia using Primer Explorer primer design software (Table 1). Reagents obtained from Lucigen for the LAMP reaction contain DNA polymerase, a reverse transcriptase enzyme, and strand displacement capabilities. This allows detection of either DNA or RNA targets. PCR master mix was prepared using these reagents (Table 2). A volume of 25µL per reaction tube was used. Each tube contained from 1-2µL of template with an effort to keep the concentration near 20ng, although concentrations ranging from 0.01ng to 100ng are acceptable. Reaction tubes were incubated at 65 °C for 25 minutes and the amplicons were immediately analyzed on an agarose gel. Control DNA from B. burgdorferi B31 ATCC 35210 and E. coli ATCC 25922 were used with each run to validate the master mix and cycling conditions. Amplicons were run on a 1.2% agarose gel using the 1 kb DNA ladder. Samples were prepared using 2µL of loading dye plus 18µL amplicon. Fifteen µL of this preparation was loaded onto the gel and run at 100 volts for 30 minutes (Figure 1).
|
Name |
Primer |
nmol/OD |
Position |
Length |
Tm |
GC% |
|
F3 |
GCTAATACCGAATAAGGTCAGT |
3.9 |
148 |
22 |
60 |
41 |
|
B3 |
CGTCATCACTTTGTCATTTCC |
4.9 |
472 |
21 |
60 |
43 |
|
FIP(F1c+F2) |
TGTGACCGTTCACCCTCTCAGCGTCTTATTAGCTAGTTGGT |
2.4 |
41 |
|||
|
BIP(B1c+B2) |
AGACTCCTACGGGAGGCAGTCTTCATTCACGCAGTGTC |
2.5 |
38 |
|||
|
LoopF |
CGGTTACTTATCATTGCCTTGG |
4.4 |
280 |
22 |
62 |
46 |
|
LoopB |
CAGCTAAGAATCTTCCGCAATG |
4.1 |
343 |
22 |
62 |
46 |
|
F2 |
GCGTCTTATTAGCTAGTTGGT |
4.5 |
224 |
21 |
60 |
43 |
|
F1c |
TGTGACCGTTCACCCTCTCA |
5.1 |
303 |
20 |
65 |
55 |
|
B2 |
TCTTCATTCACGCAGTGTC |
5.2 |
402 |
19 |
60 |
47 |
|
B1c |
AGACTCCTACGGGAGGCAG |
4.7 |
324 |
19 |
65 |
63 |
Table 1 LAMP designed primers used in the reaction mix
|
Component |
Concentration |
|
Nuclease-free water |
to 25uL/tube |
|
10X DNA Polymerase Buffer C |
1X |
|
dNTP's (25mM each) |
800µM |
|
100mM MgSO4 |
8mM |
|
5M Betaine |
0.5 M |
|
Target-specific Primer Mix |
Variable |
|
OmniAmp DNA Polymerase, 50X |
2X |
|
Template |
0.01ng - 100ng |
Table 2 All reaction mix reagents were purchased from Lucigen in the OmniAmpTM RNA/DNA Kit and the reactions were set up using the concentrations.
Figure 1 shows an example of the gel profile obtained when LAMP primers were used to amplify the DNA that codes for 16S rRNA of the Borrelia organism. The gel represents data from six patients plus a positive control, a negative control, and a water control. Gel patterns show multiple bands in a fixed configuration when positive results are obtained.
Results in Table 3 show that the LAMP PCR was positive in 53% of the positive samples. The conditions used included positive at 6 days and harvested at 6 days, positive at 6 days and harvested at 8 weeks, positive at 8 weeks and harvested at 8 weeks, DNA extraction using MagJet methodology (magnetic beads), DNA extraction using the Qiagen Kit Cat # 51306 (spin columns), Liberase TL treatment and human DNA removed, no Liberase treatment, no human DNA removed.
|
|
|
LAMP |
Plasmid |
OspC |
|
|
|
DNA extracted using Magjet |
1 |
Pos |
Neg |
Pos |
||
|
2 |
Pos |
Neg |
Pos |
Day 6 positive |
||
|
6 |
Pos |
Neg |
Neg |
Harvested Day 6 |
||
|
7 |
Pos |
Neg |
Neg |
No Liberase treatment |
||
|
8 |
Pos |
Neg |
Neg |
Human DNA not removed |
||
|
10 |
Pos |
Neg |
Pos |
|||
|
DNA extracted using MagJet |
13 |
Pos |
Neg |
Pos |
||
|
14 |
Pos |
Neg |
Neg |
Day 6 positive |
||
|
15 |
Pos |
Neg |
Neg |
Harvested 8 weeks |
||
|
16 |
Pos |
Neg |
Neg |
Treated with Liberase |
||
|
17 |
Pos |
Neg |
Neg |
Human DNA removed |
||
|
18 |
Pos |
Neg |
Neg |
|||
|
23 |
Pos |
Neg |
Pos |
|||
|
DNA exttacted using Qiagen Kit 51306 |
25 |
Pos |
Neg |
Neg |
Day 6 positive |
|
|
26 |
Pos |
Neg |
Pos |
Harvested Day 6 |
||
|
27 |
Pos |
Neg |
Neg |
Liberase treated |
||
|
28 |
Pos |
Neg |
Pos |
Human DNA removed |
||
|
57 |
Pos |
Neg |
Pos |
|||
|
DNA extracted using MagJet |
A |
Neg |
Neg |
Neg |
||
|
B |
Neg |
Neg |
Neg |
Day 6 positive |
||
|
C |
Neg |
Neg |
Pos |
Harvested Day 6 |
||
|
D |
Neg |
Neg |
Neg |
No Liberase treatment |
||
|
E |
Neg |
Neg |
Neg |
Human DNA removed |
||
|
F |
Neg |
Neg |
Neg |
Zeolite added |
||
|
DNA extracted using MP Biomedical Fast Prep extractor |
S51 |
Neg |
Neg |
ND |
||
|
S53 |
Neg |
Neg |
ND |
Day 6 positive |
||
|
S54 |
Neg |
Neg |
ND |
Harvested Day 6 |
||
|
S56 |
Neg |
Neg |
ND |
No Liberase treatment |
||
|
S64 |
Neg |
Neg |
ND |
Human DNA not removed |
||
|
MP Biomedical FastPrep 24 extractor |
8ES28 |
Pos |
ND |
ND |
8 wk pos, harvested 8 wk |
|
|
8ES45 |
Pos |
ND |
ND |
No Lib; Human removed |
||
|
DNA extracted using MagJet |
8MS1 |
Neg |
Pos |
Neg |
||
|
8MS2 |
Neg |
Neg |
Pos |
|||
|
8MS3 |
Neg |
Neg |
Pos |
8 wk positive |
||
|
8MS4 |
Neg |
Pos |
Pos |
Harvested 8 weeks |
||
|
8MS6 |
Neg |
Pos |
Neg |
No Liberase treatment |
||
|
8MS7 |
Neg |
Pos |
Neg |
Human DNA removed |
||
|
8MS9 |
Neg |
Neg |
Pos |
|||
|
8MS12 |
Neg |
Pos |
Neg |
|||
|
8MS16 |
Neg |
Pos |
Neg |
|||
|
8MS22 |
Neg |
Pos |
Pos |
|||
|
8MS24 |
Neg |
Pos |
Neg |
|||
|
DNA extracted using MagJet |
8MS61 |
Pos |
Neg |
Neg |
||
|
8MS62 |
Pos |
Neg |
Neg |
8 wk positive |
||
|
8MS64 |
Pos |
Neg |
Neg |
Harvested 8 weeks |
||
|
8MS70 |
Pos |
Neg |
Neg |
Liberase treated |
||
|
8MS73 |
Pos |
Pos |
Neg |
Human DNA removed |
||
|
25/47 |
9 of 45 |
14/40 |
||||
|
53% |
20% |
35% |
||||
Table 3 PCR results using varied harvesting timepoints, pre-processing steps, DNA extraction methods and PCR primer configurations
In twenty-two of the samples, negative results were obtained using LAMP when human DNA was not removed and/or Liberase treatment was not applied. Six samples, however, did give positive results with LAMP when pre-processing was not performed.
Data extraction and analysis can be derived from Table 3. For example, positive signal comparisons at each collection time point are made in Figure 2. Considering all pre-processing steps collectively and all PCR methods, harvesting of 8 week positives at 8 weeks gave slightly more positives than either of the other two collection times. Collective positive results showed a positive rate in 6 day positives harvested at 6 days of 29%, a positive rate in 6 day positives harvested at 8 weeks of 39%, and a positive rate in 8 week positives harvested at 8 weeks of 42%.
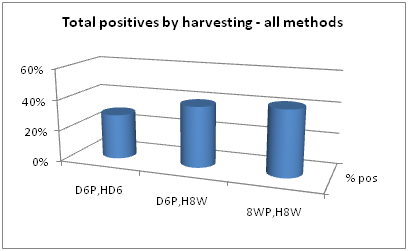
Figure 2 Collective positive rates of samples harvested at three different time points. D6P, HD6=6 day positive, harvested at 6 days. D6P, H8W=6 day positive, harvested at 8 weeks. 8WP, H8W=8 week positive, harvested at 8 weeks.
In a breakdown of the differences based on extraction method, the following comparisons are made in Figure 3. In pellets treated with Liberase TL and with human DNA removed, the LAMP targeting DNA that codes for 16S rRNA out-performed both the plasmid primers and the OspC primers when DNA extractions were performed using either MagJet or Qiagen kits.
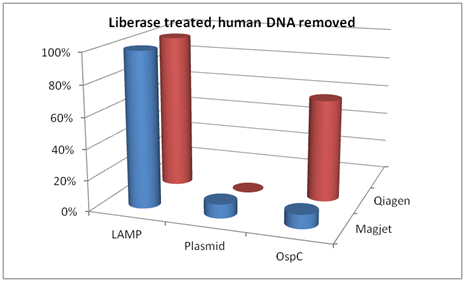
Figure 3 Comparison of LAMP 16S ribosomal PCR, plasmid PCR, and OspC PCR when DNA was extracted using either Magjet or Qiagen kit Cat #51306. These samples were treated with Liberase TL and had the human DNA removed.
Differences based on harvesting time points show additional relationships. LAMP primers performed substantially better (Figure 4) than either the plasmid primers or the OspC primers. The LAMP method showed a 100% positive rate at each harvest time point when samples were pre-treated with Liberase TL and processed to remove human DNA.
LAMP technology was originally developed around the ability to perform PCR in the field or outside of a laboratory.9-14 This has resulted in kits with workflow and simplicity advantages that can be performed outside of a biological hood on the bench in the clinical microbiology laboratory when pre-prepared lyophilized reagents are used. This application allows the addition of sample in buffer to a tube of lyophilized reagents, incubation at one temperature,15 and reads of turbidity or fluorescence using a simple instrument.16,17 An additional advantage is that the nucleic acid in the sample does not have to be extracted or purified. LAMP primers are designed to amplify in a loop formation and then prime off the loops.18,19 This serves as a way of generating considerably more product in considerably less time than conventional PCR.20
To contrast the conventional PCR process with the LAMP process, LAMP can be performed in 30 min to 2 hours from sample to signal in a single tube at one temperature at low cost. Conventional PCR from sample to signal can take anywhere from one to two workdays even when using an automated nucleic acid extractor. It requires three separate rooms in the clinical laboratory, multiple tubes, complex preparation, a multiple temperature cycling instrument, and a means to detect the amplicons that are generated. For our application, two pre-processing steps, DNA extraction, two rounds of PCR in a thermo cycler, and a gel to visualize the signal was necessary when conventional PCR techniques were used. Not only does LAMP technology reduce the time for set up and signal generation, it allows the amplification of very small amounts of DNA in the sample and overcomes inhibiting substances that interfere with conventional PCR. Our data (Figure 3 and 4) clearly shows the ability of LAMP technology to out-perform conventional PCR.
The Lucigen OmniAmp reagents, designed specifically for LAMP amplification, contain specialized nucleic acid polymerases plus reverse transcriptase and allow for the amplification of DNA and RNA. In certain situations, especially viral amplification, this is a necessary added advantage.21 A combination of LAMP primers has been used in multiplex reactions22,23and various approaches to designing the primers exist.24
Considering the various time points and processes used to prepare DNA from blood culture for the PCR process, certain aspects can be observed. For example, for the majority of positive samples, two pre-processing steps and LAMP technology for amplification gave the best results. It is interesting to note that when using simple sense and antisense primers with two rounds of PCR, standard reagents and two pre-processing steps, inconsistent results were obtained. More consistent results were displayed when LAMP primers and Lucigen OmniAmp DNA and RNA reagents were used. Lucigen reagents contain both a DNA polymerase and a reverse transcriptase. Although DNA was used in these studies, future studies will take advantage of the fact that both RNA and DNA can be amplified during the detection process.
The present study showed the pharmacological potential of the ethanolic extract of Neem bark. Our findings demonstrated that the F-EtOAc, obtained after saponification of EtCNeem, showed to be rich in phenolic and flavonoid compounds with antioxidant potential, as well as a nontoxic.
None.
Authors declare that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.