Journal of
eISSN: 2471-1381


Research Article Volume 3 Issue 3
1Department of Medicine, University of Alexandria, Egypt
2Department of Clinical and Chemical Pathology, University of Alexandria, Egypt
3Department of Pathology, University of Alexandria, Egypt
Correspondence: Hoda A El Aggan, Faculty of medicine, Al Azzareta, Midan Al Khartoum, 21521, Alexandria, Egypt, Tel 002 012 7411 7543
Received: August 12, 2016 | Published: June 30, 2017
Citation: Aggan HAE, Helmy MA, Younis LK, et al. Target of Rapamycin (TOR) and autophagy in chronic hepatitis c virus infection: relation to disease activity. J Liver Res Disord Ther. 2017;3(3):63-72. DOI: 10.15406/jlrdt.2017.03.00056
Background: In chronic hepatitis C (CHC) infection, liver disease progression results from viral persistence. Deregulation of target of rapamycin (TOR) signaling and autophagy is detected in viral infections and cancer. We studied the role of TOR and autophagy in the progression of CHC infection.
Methods: 54 patients infected with HCV, [27 with CHC; 13 with cirrhosis and 14 with hepatocellular carcinoma (HCC)]; and 15 healthy subjects were included. Quantification of serum TOR was determined using enzyme linked immunosorbent assay. Tissue samples were immune-stained using TOR and autophagy protein 5 (Atg5) polyclonal antibodies. Expression of TOR and Atg5 was scored semi-quantitatively.
Results: Expression and serum TOR were higher in HCC patients compared to patients without HCC (P<0.05). Serum and expression of TOR were positively correlated to inflammation, fibrosis, steatosis and tumor characteristics (Alpha-fetoprotein, maximum diameter, tumor grade and CLIP stage) (P<0.05). Expression of Atg5 was inversely correlated with inflammation, fibrosis and tumor characteristics (P<0.05), Atg5 showed significant inverse correlation with serum and intra-hepatic expression of TOR (P<0.05). Serum TOR showed high sensitivity and specificity in discriminating infected patients with and without HCC at a cut-off value of 4.55ng/ml [Area under ROC curve=0.970].
Conclusion: Activation of TOR plays an important role in progression of HCV related liver disease possibly though autophagy suppression and they could be a potential therapeutic target. Serum TOR is a potential seromarker in discriminating HCV infected patients with and without HCC with high sensitivity and specificity.
Keywords: chronic liver diseases, liver cirrhosis, hepatocyte necrosis, rapamycin, autophagy
HCV, hepatitis c virus; CHC, chronic hepatitis C; TOR, target of rapamycin; HCC, hepatocellular carcinoma; Atg5, autophagy protein 5; AST, aspartate transaminase; ALT, alanine transaminase; GGT, gamma glutamyl transpeptidase; PA, prothrombin activity; INR, international normalized ratio; PCR, polymerase chain reaction; AFP, alfa fetoprotein; MELD, model for end stage liver disease; BCLC, barcelona clinic liver cancer; CLIP, cancer of the liver italian program; IHC, immuno histo chemical; ROC, receiver-operating characteristic
Hepatitis C virus (HCV) is a well-established cause of wide range of chronic liver diseases.1 Because acute HCV infection is rarely cleared, most patients develop chronic hepatitis.2 During the course of chronic HCV infection, inflammatory milieu, hepatocyte necrosis, steatosis, oxidative stress, and progressive liver fibrosis of variable degrees, eventually result in irreversible cirrhosis and hepatocellular carcinoma (HCC).3 The oncogenic process of HCV infection probably requires multiple steps of genetic and epigenetic alterations and the activation of cellular oncogenic.4,5 Viral structural and non-structural proteins are accused to orchestrate several hepatocyte signaling pathways with subsequent interference with cellular biological activities.6 Although the course of HCV infection is influenced by complex host-virus interplay, yet, the potential mechanism(s) underlying viral persistence and disease progression are not fully understood.7,8 Intensive research to unravel the multifunctional molecular pathway(s) implicated in the progression of chronic HCV infection could identify novel therapeutic targets to tackle this devastating disease.9 Among cell signaling pathways hijacked by many viral infections, mammalian target of rapamycin (mTOR) signaling has been extensively studied. The mTOR is a 289-kDa serine/threonine protein kinase, a member the phosphoinositol 3-kinase (PI3K)-related kinase family.10The mTOR signaling is activated when the genetic and environmental milieu is optimal for cellular growth, and diminishes under stressful conditions including insufficient nutrients, energy depletion, DNA damage and hypoxia.11,12 The mTOR signaling integrates intracellular and extracellular signals and serves as a central regulator of cell metabolism, growth, proliferation and survival.13 In addition, mTOR is a key player in the innate and adaptive immune responses.14In recent years, increasing evidence demonstrates that mTOR is down-regulator of the intracellular process of autophagy in response to cellular physiological conditions and environmental stress.15,16Autophagy or ‘self-eating’ is characterized by the formation of double-membrane vesicles, known as Autophagosome, which sequester cellular constituents and deliver them to the lysosomes.17Autophagy involves cascade of events including a series of complexes composed of autophagy proteins (Atg) that assemble Autophagosome.18Autophagy can be induced by a variety of stimuli (e.g. nutrient deprivation, hypoxia, cytokines, hormones, viruses and DNA damage).19 Beyond maintaining cellular homeostasis,20 autophagy is involved in multiple biological processes including cell quality control, energetic balance, remodeling, and defense against extracellular insults and pathogens.21 It is likely that dysregulation of the mTOR-autophagy pathway may contribute too many human disorders including viral infections.22Inflammatory disorders23,24and cancer25,26and could be an attractive avenue for future therapeutic approaches.16 The aim of the present work was to study the role of mTOR and autophagy in the progression of HCV-related liver disease.
The study included 54 treatment naïve patients with seropositivity of HCV antibodies, HCV-RNA level and histopathological findings consistent with chronic HCV infection. Patients were subdivided into 27 patients with chronic hepatitis C (CHC), 13 cirrhotic patients without HCC and 14 patients with HCC who underwent surgical resection of the tumor]. Exclusion criteria included seropositivity for hepatitis B virus; alcohol consumption; concomitant schistosomiasis; hepatic decompensation, bleeding diathesis; diabetes mellitus, connective tissue diseases or other autoimmune diseases, infections, malignancy, and cardiac, respiratory or renal disease, previous antiviral treatment or locoregional or systemic therapy for HCC. At the same time, 15 age- and sex-matched healthy subjects with no evidence of liver disease were included in the study as a control group to obtain the normal range of biochemical assays. The study was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines. An informed consent was obtained from all subjects included in the study. On admission, the following clinic-laboratory data were recorded: Age, sex, the apparent duration and possible risk factors of HCV infection, symptoms and signs of chronic liver disease, liver and spleen size, presence of ascites, serum aspartate transaminase (AST), serum Alanine transaminase (ALT), albumin, bilirubin and gamma glutamyl transpeptidase (GGT), and prothrombin activity (PA)/international normalized ratio (INR), serum creatinine, serum HCV antibodies, hepatitis B surface antigen, hepatitis B core antibody and serum HCV RNA levels using real time polymerase chain reaction (PCR) and serum Alfa fetoprotein (AFP). Ultrasonographic and triphasic CT examination was performed for assessment of liver and spleen, presence of ascites and tumor characteristics in patients with HCC. The severity of liver disease in cirrhotic patients with and without HCC was graded according to Child-Pugh classification and the Model for End Stage Liver Disease (MELD) score. The staging of HCC was determined according to the Barcelona Clinic Liver Cancer (BCLC) and the Cancer of the Liver Italian Program (CLIP).
Serum mTOR levels measurement
The mTOR protein levels in sera of patients and healthy subjects were measured using sandwich ELISA technique using human solid phase sandwich ELISA kit (Human mTOR ELISA kit, MBS2505637, My Bio Source, Inc., CA, USA) according to the manufacturer’s instructions.
Histopathological examination
Liver biopsy was performed in patients with CHC as well as representative samples of surgically-resected HCCs and surrounding non-neoplastic tissues were also obtained for histopathological and immunohistochemical (IHC) assessment. Formalin fixed paraffin embedded tissue specimens were stained with hematoxylin and eosin stain for routine histopathological examination to assess the histological activity grade and fibrosis stage according to the METAVIR scoring system and steatosis grade for liver specimens from 30 patients with chronic HCV infection in absence of HCC and the surrounding non-neoplastic tissues. The diagnosis of HCC was confirmed by histopathological examination and the tumor was graded according to Edmonson and Steiner's criteria.27
Immunohistochemical staining
Immunohistochemical staining was performed using Ultravision Avidin-Biotin Detection System (Thermo Scientific, CA, USA). The tissue sections (5μm-thick) were placed on amino-propyl-tri-ethoxy-silane coated glass slides, dried and de-waxed in xylene and ethanol over 15-20 minutes, then rehydrated through a series of graded alcohol to water. Endogenous peroxidase activity was blocked by incubation in two changes of 3% (v/v) hydrogen peroxide in phosphate buffered saline (PBS) (pH 7.4) for 10 minutes at room temperature. Antigen retrieval was done by heating slides in 10 mM sodium citrate buffer, pH 6.0 at 95 -100°C for 10 minutes then removed from heat and let stand at room temperature in buffer for 20 minutes then rinsed in Tris-buffered saline for 1 minute. Tissue sections were incubated with primary antibodies [rabbit anti-human mTOR polyclonal antibody (1:30, MBS301883, My Biosource, Inc., CA, USA) and rabbit Anti-human autophagy related peptide-5 (Atg5) polyclonal antibody (1:500, MBS150643, My Bio source, Inc., CA, USA)] for 30 minutes at room temperature. Slides were rinsed with PBS for 5 min, and nonspecific antibody binding was blocked by incubation of the tissue slides with horse serum before proceeding to the primary antibodies. Immunostaining was visualized using DAB (3, 3’ Diamono-benzidine-tetrahydrocholride) chromogen (Dako, Denmark). Positive staining was recognized under light microscope as a diffuse cytoplasmic and/or nuclear brown color stain. Sections were counterstained with Mayer’s hematoxylin solution. Replacement of the primary antibody with rabbit IgG was used as negative control in each batch of experiment and was run in parallel.
Evaluation of mTOR and Atg5 immunostaining
Immunostaining was evaluated semi-quantitatively according to the percentage of positively-stained cells in non-overlapping microscopic fields. The percentage of positively stained cells was determined by counting a minimum of 100 cells, and the average was recorded, and scored as follows: (0)=absent staining; (1)=weak, <10% of cells were positive; (2)=moderate, 10-50% of cells were positive or (3) = strong, >50% of cells were positive.28,29
Statistical analysis
We used the Statistical Package for Social Sciences (SPSS version 20.0) software. The data were expressed as mean ± SD or proportions. Comparison between two means was performed using the non-parametric Mann-Whitney U-test for abnormally distributed quantitative variables. The one-way ANOVA test was used for comparing the three groups with post hoc comparisons. Comparison between proportions was determined by the Chi square (c2) test or Fisher’s Exact test (FET). Correlations between variables were analyzed by using Pearson's correlation coefficient or Spearman’s rank test as appropriate. Statistical significance was assessed at P<0.05. All calculated P values were two-tailed. The sensitivity and specificity of serum mTOR in discriminating cirrhotic patients with and without HCC were assessed by plotting a receiver-operating characteristic (ROC) curve and determining its cut-off value.
Demographic and laboratory data of study groups are summarized in Table 1.
Parameters |
CHC Patients (n = 27) |
Cirrhosis Without HCC (n = 13) |
HCC (n = 14) |
Age (years) |
|||
Range |
20-53 |
41-60 |
35-69 |
Mean ± SD |
39.70 ± 9.97 |
51.08 ± 6.21 |
55.43 ± 9.52 |
Sex (%) |
|||
Male |
12 (44,4) |
10 (76.9) |
9 (64.3) |
Female |
15 (55.6) |
3 (23.1) |
5 (35.7) |
AST(U/L) |
8-80 (37.81±14.77) |
23-95 (60.46± 22.87) |
15-92 (54.86±22.97) |
ALT(U/L) |
9-140 (52.44±29.73) |
24-163 (68.85±35.84) |
25-198 (66.29±40.53) |
GGT (U/L) |
12.2-49.7 (25.82 ± 8.05) |
18.6-45.2 (32.16± 9.08) |
18.9-29.3 (25.41±3.23) |
Serum AFP (ng/ml) |
1.2-23.5 (7.64±5.27) |
3.6-88 (33.86±26.18) |
11-20973 (2436.86±5706.94) |
HCV - RNA(x103 IU/ml) |
19-6500 (923.27±1504.43) |
52.6-6231 (1543.53±2208.21) |
110-2300 (675.00±626.42) |
Child-Pugh score |
- |
5-7 (6.08 ± 0.64) |
5-7 (5.71±0.73) |
MELD |
- |
6-9 (7.62±0.87) |
6-10 (8.14±1.46) |
Table 1 Demographic, Clinical-laboratory characteristics of patients with chronic hepatitis C (CHC) and cirrhosis with and without hepatocellular carcinoma (HCC).
Serum mTOR levels
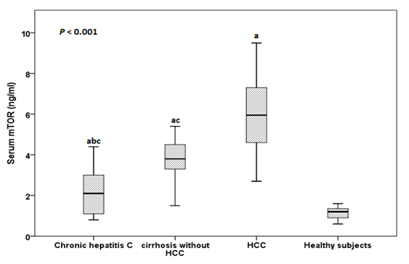
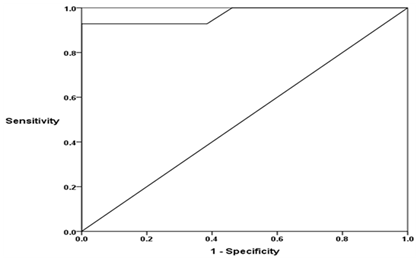
The serum mTOR levels ranged between 0.8-4.4 ng/ml in patients with CHC, between 1.5-4.5 ng/ml in cirrhosis without HCC patients, between 3.7-9.5 ng/ml in patients with HCC and between 0.6-1.6 ng/ml in healthy subjects. The mean serum mTOR levels was significantly higher in patients with CHC, patients with cirrhosis without HCC and HCC than in healthy subjects (2.22±1.20 ng/ml, 3.49±0.79 ng/ml and 6.19±1.63 ng/ml vs 1.13±0.30 ng/ml respectively), in patients with cirrhosis and HCC than in patients with CHC and in patients with HCC than in patients with cirrhosis (P<0.001) (Figure 1). By plotting Receiver operating characteristic (ROC) curve, the sensitivity and specificity of serum mTOR levels in discriminating cirrhotic patients with and without HCC were 92.9% and 100% respectively at a cut-off value of 4.55 ng/ml (AUC = 0.970), (Figure 2).
Histopathological examination
In HCV infected patients without HCC and according to METAVIR scoring system, patients with chronic HCV infection showed histological activity grade A1 in 10 (33.3%) patients, A2 in 13 (43.3%) patients and A3 in 7 (23.3%) patients while fibrosis stage was classified as F1 in 7 (23.3%) patients, F2 in 15 (50.0%) patients, F3 in 5 (16.7%) patients and F4 in 3 (10%) patients. Steatosis was absent in 5 (16.7%) patients, mild in 10 (33.3%) patients, moderate in 8 (26.7%) patients and marked in 7 (23.3%) patients. Grossly; in HCC patients, the maximum diameter of the tumor ranged between 2.7 and 11.6 cm with a mean value of 6.19 ± 2.93 cm. The tumors were uni-nodular in all patients and were located in the right lobe in 5 (35.7 %) patients and in the left lobe in 9 (64.3%) patients. The extension of all surgically-resected HCCs was <50% of the liver. According to Edmondson and Steiner’s grading system, HCCs were graded as grade II in 7 (50.0%) patients, grade III in 2 (14.3%) patients and grade IV in 5 (35.7%) patients. The surrounding non-neoplastic liver tissues showed cirrhosis (METAVIR F4) in all patients with HCC. The histological activity grade was A2 in 4 (28.6%) patients and A3 in 10 (71.4%) patients. Steatosis was mild in 2 (14.3%) patients, moderate in 11 (78.6%) patients and marked in one (7.1%) patient.
Immunostaining for mTOR
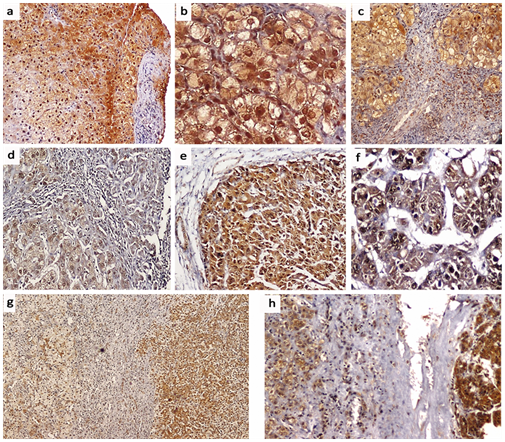
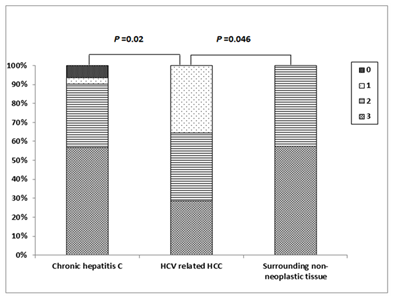
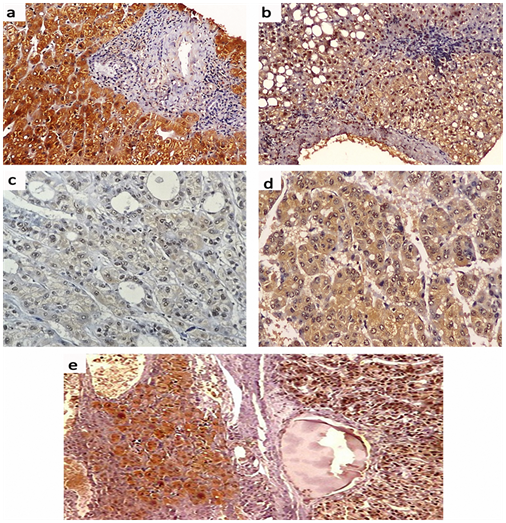
Positive Immunostaining for mTOR was detectable as cytoplasmic and/or nuclear staining in 18 (60.6%) of patients with chronic HCV infection; of them, 4 (13.3%) patients showed weak staining, 6 (20.0%) patients showed moderate staining and 8 (26.7%) patients showed strong staining. In HCV-related HCCs, positive mTOR Immunostaining was detected in 12 (85.7%) patients; of them, weak staining was found in one (7.1%) patient, moderate staining in one (7.1%) patient and strong staining in 10 (71.4%) patients. The surrounding non-neoplastic liver tissues showed positive mTOR Immunostaining in 12 (85.7%) patients; among them, weak staining in 3 (21.4%) patients, moderate staining in 6 (42.9%) patients and strong staining in 3 (21.4%) patients (Figures 3 & 4). There was a significant difference in the distribution of mTOR expression among different groups (FET=13.096, P=0.03). The mTOR expression was significantly higher in HCV-related HCCs than in chronic HCV infection tissues (FET=7.201, P=0.049) and the surrounding non-neoplastic liver tissues (FET=8.149, P=0.035) while there was no significant difference between the latter two groups (FET=4.321, P=0.235).

Immunostaining for Atg5
Cytoplasmic positive Immunostaining for Atg5 was detectable in 28 (93.3%) patients with chronic HCV infection; one (3.3%) patient showed weak staining, 10 (33.3%) patients showed moderate staining and 17 (56.7%) patients showed strong staining. In HCV-related HCCs, positive Atg5 Immunostaining was detected in all patients; weak staining in 5 (35.7%) patient, moderate staining in 5 (35.7%) patient and strong staining in 4 (28.6%) patients while the surrounding non-neoplastic liver tissues showed moderate Atg5 Immunostaining in 6 (42.9%) patients and strong staining in 8 (57.1%) patients (Figures 5 & 6). There was a significant difference in the distribution of Atg5 expression among different groups (FET=11.297, P = 0.038). Atg5 expression was significantly higher in HCV-related HCCs than in the chronic HCV infection tissues (FET=8.561, P=0.02) and the surrounding non-neoplastic liver tissues (FET = 6.271, P=0.046) while there was no significant difference between the latter two groups (FET = 1.342, P=0.840).
Statistical correlations in HCV infected patients without HCC
The serum mTOR levels and intra-hepatic mTOR expression showed significant positive correlations with serum AST levels (r=0.547, P<0.001 and r=0.555, P=0.001 respectively), serum ALT levels (r=0.624, P< 0.001 and r=0.501, P=0.005 respectively), the METAVIR histological activity grade (r=0.379, P=0.039 and r=0.442, P=0.015 respectively) and fibrosis stage (r=0.547, P=0.002 and r=0.489, P=0.006 respectively) and steatosis grade (r=0.610, P<0.001 and r=0.393, P=0.032 respectively). The intra-hepatic Atg5 expression showed significant inverse correlations with serum AST levels (r= - 0.409, P = 0.025), serum ALT levels (r= -0.421, P= 0.021) and the METAVIR histological activity grade (r= -0.595, P=0.001) and fibrosis stage (r= -0.420, P= 0.021). There was no significant correlation with steatosis grade (P> 0.05). Serum mTOR levels and intra-hepatic mTOR expression were positively correlated (r=0.507, P=0.004) and both showed inverse correlations with the intra-hepatic Atg5 expression (r= -0.398, P= 0.029 and r = -0.611, P< 0.001 respectively) (Table 2).
Parameters |
Serum mTOR (ng/ml) (n = 40) |
mTOR Expression(n = 30) |
Atg5 Expression (n = 30) |
|||
|---|---|---|---|---|---|---|
r |
P |
r * |
P |
r * |
P value |
|
Age (years) |
0.194 |
0.232 |
0.029 |
0.88 |
-0.045 |
0.815 |
Serum AST (U/L) |
0.547 |
< 0.001 |
0.555 |
0.001 |
-0.409 |
0.025 |
Serum ALT (U/L) |
0.624 |
< 0.001 |
0.501 |
0.005 |
-0.421 |
0.021 |
Serum GGT (U/L) |
0.232 |
0.151 |
0.214 |
0.255 |
-0.017 |
0.93 |
Serum HCV-RNA (x103 IU/ml) |
0.091 |
0.575 |
-0.272 |
0.146 |
0.324 |
0.081 |
METAVIR: (n = 30) |
||||||
- Histological activity grade* |
0.379 |
0.039 |
0.442 |
0.015 |
-0.595 |
0.001 |
- Fibrosis stage* |
0.547 |
0.002 |
0.489 |
0.006 |
-0.42 |
0.021 |
Steatosis grade* (n = 30) |
0.61 |
< 0.001 |
0.393 |
0.032 |
-0.19 |
0.313 |
mTOR expression* (n = 30) |
0.507 |
0.004 |
- |
- |
- |
- |
Atg5 expression* (n = 30) |
-0.398 |
0.029 |
-0.611 |
< 0.001 |
- |
- |
Table 2 Statistical correlations (“r” value) between serum mammalian target of rapamycin (mTOR) levels, intra-hepatic expression of mTOR and autophagy-related protein 5 (Atg5) and other parameters in patients with chronic HCV infection without hepatocellular carcinoma.
*Spearman rho correlation
Statistical correlations in patients with HCC
The serum mTOR levels and mTOR expression in HCCs were positively correlated (r= 0.571, P= 0.033) and both showed inverse correlations with the Atg5 expression in HCCs (r= -0.703, P= 0.005 and r= -0.818, P < 0.001 respectively). Serum mTOR levels and mTOR expression in HCCs showed significant positive correlations with serum AFP levels (r= 0.725, P= 0.003 and r= 0.728, P= 0.003 respectively), HCC maximum diameter (r= 0.618, P= 0.018 and r= 0.574, P= 0.032 respectively), CLIP stage (r= 0.537, P= 0.032 and r= 0.762, P= 0.002 respectively) and HCC histological grade (r= 0.736, P= 0.003 and r= 0.593, P= 0.025 respectively). The Atg5 expression in HCCs showed significant inverse correlations with serum AFP (r= -0.713, P= 0.004), CLIP stage (r= -0.836, P< 0.001), HCC maximum diameter (r= -0.840, P< 0.001), BCLC stage (P= 0.002) and HCC histological grade (r= -0.808, P< 0.001), (Table 3).
Parameters |
Serum mTOR (ng/ml) |
mTOR Expression |
Atg5 Expression |
|||
|---|---|---|---|---|---|---|
r value |
P value |
r *value |
P value |
r *value |
P value |
|
Serum AFP (ng/ml) |
0.725 |
0.003 |
0. 728 |
0.003 |
-0.713 |
0.004 |
Child-Pugh score* |
0.11 |
0.707 |
-0.048 |
0.87 |
-0.107 |
0.717 |
MELD score* |
0.297 |
0.303 |
0.136 |
0.643 |
-0.199 |
0.495 |
HCC maximum diameter (cm) |
0.618 |
0.018 |
0.574 |
0.032 |
-0.84 |
<0.001 |
CLIP stage* |
0.573 |
0.032 |
0.762 |
0.002 |
-0.836 |
< 0.001 |
HCC grade* |
0.736 |
0.003 |
0.593 |
0.025 |
-0.808 |
< 0.001 |
mTOR expression* |
0.571 |
0.033 |
- |
- |
- |
- |
Atg5 expression* |
-0.703 |
0.005 |
-0.818 |
< 0.001 |
- |
- |
Surrounding Non-Neoplastic Liver Tissue |
||||||
mTOR expression* |
0.119 |
0.684 |
0.318 |
0.268 |
-0.134 |
0.649 |
Atg5 expression* |
0.395 |
0.162 |
0.112 |
0.702 |
-0.266 |
0.359 |
Table 3 Statistical correlations (“r” value) between serum mammalian target of rapamycin (mTOR) levels, intra-hepatic expression of mTOR and autophagy-related protein 5 (Atg5) and other parameters in patients with hepatocellular carcinoma (HCC) (n = 14).
*Spearman rho correlation.
In the present study, serum mTOR levels were significantly higher in CHC patients than in healthy subjects and was associated and positively correlated with intra-hepatic mTOR expression in 60% of patients. This suggests that mTOR was activated in chronic HCV infection. In previous studies, expression of total and phosphorylated-mTOR and its downstream substrates was increased in HCV-infected Huh 7.5cells.30‒32A recent study has also shown that HCV NS5A activated mTOR and its substrates under serum-starved conditions in Huh7 and HCV subgenomic replicon cells.33 NS5A knockdown abrogated phosphorylation of S6K1 and 4E-BP1 suggesting that NS5A specifically induced mTOR activity.33Activation of mTOR by HCV may enable the virus to orchestrate cell activities for the virus own benefit leading to HCV persistence and chronicity.33 Through achieving significant viral protein translation and lipid production needed for virus assembly; leading to a steady-state of replication.34 In the present study, the serum mTOR levels and intra-hepatic mTOR staining score were positively correlated with serum ALT, AST and the METAVIR histological activity grade and fibrosis stage in patients without HCC suggesting an association of mTOR activation with inflammation and fibrogenesis. Also, there was a progressive increase in serum mTOR levels from CHC to cirrhosis. Activation of mTOR has been reported in other experimental inflammatory liver diseases such as fatty liver disease35 and immune-mediated hepatitis.36 Artificially induced hepatitis in experimental mice or hepatoblastoma HepG2 cells, led to mTORC1 activation.37 Also, in genetic mouse model, liver-specific knockout of tuberous sclerosis 1 (TSC1), a negative regulator of mTOR, was associated with increased ALT and AST, focal necrosis and inflammation with macrophage infiltration.38 In rats with cirrhotic portal hypertension induced by bile duct ligation, mTOR was markedly activated and treatment with mTOR inhibitors decreased ALT, AST, alkaline phosphatase, intra-hepatic neutrophils and lymphocytes and reduced TNF-α and inducible nitric oxide synthase mRNA expression. Moreover, reduction of fibrosis up to 70%, numbers of cholangiocytes and myo fibroblasts, and hepatic extracellular matrix deposition was observed.39,40 Moreover, in post liver transplant patients, the use of mTOR inhibitors was associated with slower rate of liver fibrosis.41
The relationship between mTOR and inflammation could be mutual where inflammatory cytokines such as TNF-α may activate mTOR42 and mTOR promotes the expression of pro-inflammatory cytokines like TNF-α, IL-6, IL-1α and IL-1b.35,39,40 In addition, mTOR activation enhances the expression of monocyte chemotactic protein-l and hepatic macrophage infiltration and polarization with M2 to M1 phenotype switch.43mTOR is suggested to promote fibrogenesis through promotion of proliferation and trans-differentiation of HSCs into myo fibroblast-like cells.44Also, recent studies linked mTOR to the signaling pathway of the αVβ3 integrin, a receptor for vitronectin, which is the primary molecule required for fibroblast attachment and spreading.45Another possible mechanism that promotes liver fibrosis progression is the development of hepatic steatosis.46 The present study showed that serum mTOR levels and intra-hepatic mTOR expression were positively correlated with steatosis grade in HCV infected patients without HCC suggesting a possible role of mTOR in hepatic lipid accumulation. Previous studies demonstrated that mTOR gene and protein expression were up-regulated in patients with fatty liver diseases; while rapamycin prevented the development of hepatic steatosis.47,48 Moreover, rapamycin reduced hepatic uptake of free fatty acids and triglycerides with alleviation of hepatic steatosis in hepatoblastoma HepG2 cells.49 In infected cells, lipid droplets facilitate HCV core protein storage and virus assembly.50 At the molecular level, mTOR was found to regulate lipogenesis through negative regulation of lipin-1 and/or activation of S6K.51 Moreover; mTOR signaling up-regulates PPARγ with promotion of fatty acid uptake, synthesis, esterification, and storage.52
Also, in the current study, the development of HCC was associated with a further significant increase in both serum levels and intra-tumoral mTOR staining which was significantly higher than the surrounding non-neoplastic tissues, and positively correlated with serum AFP levels, tumor size, Edmonson grade and CLIP stage. These findings indicate that mTOR activation progressed from chronic hepatitis to cirrhosis to HCC with association with aggressive tumor behavior. In agreement, expression of mTOR protein as well as mRNA was detected in high percent of HCC cases in contrast to adjacent non-cancerous liver tissues.53‒56 Similarly, an incremental activation of mTOR was detected in HCC, dysplastic nodules and non-neoplastic surrounding tissues when compared with normal livers, with the highest expression being in HCC.57,58 In HCCs, activation of the mTOR signaling has been associated with poor differentiation, high TNM and BCLC staging, high AFP, intra-hepatic metastasis, vascular invasion, angiogenesis, high proliferation index and post-transplant recurrence; whereas inhibition of mTOR could suppress tumor growth and sensitize tumor cell to chemotherapy or other targeted therapies.53,54,56,57,59‒62 In several preclinical studies, liver-specific TSC1 knockout mice showed constitutively elevated mTOR signaling and developed sporadic HCC.38,55,63,64 Besides the prognostic potential of mTOR, the present study showed that serum mTOR levels could be of diagnostic value for the development of HCC in patients infected with HCV with sensitivity and specificity of 92.2% and 100% respectively at a cut-off value 4.55ng/ml. To our knowledge, this is the first study to provide data on serum levels of mTOR in patients with HCV-related liver disease and we suggest serum mTOR as a potential sero-marker to discriminate cirrhotic patients with and without HCC; a finding that should be verified in a large number of patients. The mTOR pathway promotes cell proliferation and survival by inhibiting apoptosis and enhancing progression from the G1 to the S phase, leading to accumulation of mutations and tumor development.65 Moreover, mTOR signaling represents a central avenue for several oncogenic pathways and aberrant activation of mTOR in the majority of human cancers often occurs as a result of dysregulation of a signaling network of oncogenes and tumor suppressors lying upstream of mTOR.66 Activation of mTOR accelerates cell growth through promoting protein synthesis67 as well as enhancing angiogenesis by activating VEGF signaling and expression of nitric oxide and angiopoietins during hypoxia.56,68 Moreover, mTOR related carcinogenesis may be mediated through its controlling effect over the intracellular stress response processes including autophagy.15 The present study showed that Atg5 was detectable in hepatocyte of most of HCV infected patients without HCC with significant inverse correlation with serum amino transferases and the METAVIR histological activity grade and fibrosis stage while there was no correlation to steatosis. In addition, the expression of Atg5 was significantly lower in HCCs than cirrhotic and the surrounding non-neoplastic liver tissues and was inversely correlated with serum AFP levels, tumor size, histological grade and stage. These findings suggest that autophagy is a tumor suppressor and a down regulated autophagy may contribute to the development and progression of HCC.
Limited studies with similar results evaluated autophagy in patients with chronic HCV infection.69,70 However, in experimental studies autophagy was activated in HCV-RNA expressing cultured cells.71‒74 It has been proposed that HCV induces autophagy through ER stress and the unfolded protein response.71 However, whether HCV induces a functional or incomplete autophagy process remains controversial. Emerging lines of evidence suggested that HCV stimulates an incomplete autophagic response.71,75 HCV might usurp the autophagy pathway to use it as a mechanism for initiating HCV RNA replication and thereafter, it might inhibit auto lysosome maturation to protect itself from xenophagic degradation.71,76 Autophagy facilitates inflammation resolution through suppression of inflammasome activation,77 removal of apoptotic corpses78 and cell protection from oxidative stress through mitophagy.79 This could explain our results as well as similar previous reports.79‒82 The role of autophagy in the liver fibrosis is controversial. Autophagy may promote liver fibrosis by enhancing HSC trans-differentiation through lipophagy83 which provide free fatty acids to fuel HSC activation.84 It is therefore possible that selective inhibition of autophagy in liver fibrogenic cells might be used to treat patients with liver fibrosis. On the other hand, anti-fibrosis role of autophagy has been suggested. Increased renal collagen deposition and fibrosis was observed in Beclin 1 heterozygous deletion mice, suggesting autophagy may suppress fibrosis.85 Therefore, further studies are needed to clarify the role of autophagy in the process of liver fibrosis before a therapeutic approach targeting autophagy can be used. Previous studies showed that the mRNA levels of Beclin 1 were lower in HCC tissues than in chronic hepatitis or cirrhosis.86,87In addition, other in vivo and in vitro studies supported that the autophagic pathway was inhibited in HCC.71,72,88‒90 The role of autophagy in hepato-carcinogenesis remains controversial. Dual effects of autophagy in cancer have been proposed depending on the tumor microenvironment.25,91 Autophagy may function as a tumor suppressor through removal of damaged and senescent mitochondria and thus, restricts oxidative and metabolic stress and limits genetic instability and oncogenic mutations.25,92 On the other hand, mounting evidence suggests that autophagy may have cancer promoting effect91through providing stress tolerance, and inhibition of apoptosis.93 These controversies are important and needs further studies to clarify because many current cancer therapy agents activate autophagy.94 We observed that Atg5 expression was inversely correlated with serum levels and intra-hepatic expression of mTOR. In agreement, knockdown of mTOR or rapamycin treatment significantly enhanced autophagy in HepG2 cells suggesting that autophagy is induced by repressing the mTOR.88 These observations denote that mTOR may utilize autophagy inhibition to augment inflammation, fibrosis and cancer cell progression and survival.16However, the lack of a correlation between autophagy and hepatic steatosis in the present study may suggest that mTOR-induced lipogenesis was not mediated by autophagy suppression. Meanwhile, simultaneous high expression (score 2 or 3) of both mTOR and Atg5 was detected in 44.8% patients. This unexpected finding indicates that autophagy was induced despite mTOR activation. Previous studies found that autophagy was up-regulated in HCV-infected hepatocytes with increased expression of mTOR and vice versa.30,95This observation is explained by finding that autophagy induction may be mediated by the class III PI3K, Vps34, which is also known to be involved in activation of mTOR.96 Thus, mTOR signaling and autophagy may be inversely inter-related or may act concurrently in HCV infection. A potential explanation of this paradox is likely that HCV infection induces autophagy for establishment of infection, while activates mTOR signaling for hepatocyte growth.30
The mTOR is activated in the course of chronic HCV infection. Activation of mTOR as well as deregulated autophagy play an important role in progression of HCV related liver disease and they could be a potential therapeutic target. Also, serum mTOR is a potential seromarker in discriminating HCV infected patients with and without HCC with high sensitivity and specificity.
None.
Author declares that there is no conflict of interest.

©2017 Aggan, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.