Journal of
eISSN: 2377-4312


Research Article Volume 13 Issue 2
1Department of Animal Production, Adamawa State University, Mubi, Adamawa State, Nigeria
2Department of Animal Science, Ahmadu Bello University, Zaria, Kaduna State, Nigeria
3 Department of Livestock and Fisheries, National Agricultural Extension and Research Liaison Services, Zaria, Kaduna State, Nigeria
4Department of Animal Production and Health Technology, Federal Polytechnic, Mubi
5Department of Animal Science, School of Agriculture, University of Cape Coast, Cape Coast, Ghana
6Hungarian University of Agriculture and Life Sciences, MATE - Kaposvar Campus, Guba Sandor Utca 40.7400, Kaposvar, Hungary
Correspondence:
Received: July 30, 2024 | Published: August 19, 2024
Citation: Abbaya HY, Kabir M, Iyiola-Tunji AO, et al. Genetic studies of heat shock protein (HSP90AA1) gene in some Nigerian indigenous cattle. J Dairy Vet Anim Res. 2024;13(2):76-81. DOI: 10.15406/jdvar.2024.13.00353
The experiment was carried out to study the single nucleotide polymorphism of Heat Shock Protein (HSP90AA1) gene in selected Nigerian indigenous cattle in Adamawa State. Blood taken on eighty (80) lactating animals within their early lactation (1-60). HSP90AA1 gene was extracted and sequenced. Eight (8) sequences were generated from the selected breeds and were deposited in the GenBank with accession numbers MZ2355888 – MZ2355895. The sequences generated in this study revealed six (6) polymorphic sites in the coding regions (136 G>A, 136 G>A, 89 C>G, 89 C>G, 86 A>G and 86 A>G) that defined four haplotypes. Analysis of Molecular Variance (AMOVA) of the four breeds revealed that 58.18% of the variation was among breeds than within breeds (41.81%). It was concluded that there is more genetic variation among the studied breeds than within the breeds for HSP90AA1 gene.
Keywords: cattle, disease, animals, genetic
One strategy for reducing the magnitude of heat stress is to select animals that are genetically resistant to heat stress.1 The goal of any selection program for heat tolerance must be to develop cattle that can perform in challenging environments while maintaining high levels of productivity and carcass performance.2 Tropically adapted cattle are known for their ability to tolerate heat stress while maintaining standards of milk yield, reproduction, and disease resistance.3
The importance of thermo-tolerance in dairy cattle prompted genetic investigations of some genes that serve as candidate genes for heat tolerance in dairy cattle. One of these genes is Heat Shock Protein gene.4 Heat Shock Proteins (HSPs) are group of proteins which are synthesized during heat stress and few studies have shown an association between Single Nucleotide Polymorphism at HSP90AA1 genes and heat resistance in different species. It was reported that organisms respond to environmental stress through reprograming leading to the production of heat shock proteins (HSPs).5
The genetic variation in HSP genes among breeds and the central role that HSPA1A has in coordinating thermal tolerance suggest that HSP90AA1 is a candidate gene for identification of genetic markers and that there is an opportunity to improve thermal tolerance through marker-assisted selection.6 HSP90AA1gene in Deoni cattle (Bos indicus) has been found to be polymorphic and showed significant association with productive and reproductive parameters.7
Many earlier studies have shown an association between single nucleotide polymorphisms (SNPs) at certain HSP genes and heat resistance.8,9 HSP genes have been reported to be associated with heat tolerance and reproduction performance in cattle. Studies as molecular characterization based on Single Nucleotide Polymorphism (SNP) has become a tool in identifying genetic markers that allow the detection of variations between populations as well as detecting various regions of the DNA that can account for variations and relationships among populations.10,11
Thermo-tolerance appears to be a quantitative trait influenced by many regions of the genome, and genomic studies have identified regions of the genome that appear to be important for regulation of body temperature in both beef and dairy cattle.11 The aim of this paper therefore is to explore the genetic variations in Heat Shock Protein (HSP90AA1) that exist in Adamawa Gudali, Rahaji, Bolokoji and Bunaji breeds of indigenous cattle in Nigeria.
The study was conducted on four selected herds in Adamawa State (Mubi, Hong, Gombi and Song Local Government Areas). Adamawa State is located at an altitude of 200 to 300 meters above sea level, between latitude 9º20’ and 9º33’N and longitude 120 30’ and 12º50’ E (Adebayo et al., 2020). It has average daily minimum and maximum temperatures of 23.2 and 35.2ºC respectively. The average annual rainfall is 718.1 millimetres and relative humidity, 44.2 %. It occupies an area of 39,742.12 square kilometers.12
Eighty (80) clinically healthy dairy cows made up of twenty (20) each of Adamawa Gudali (AG), Rahaji (RJ) also known as Red Bororo, Bokoloji (BK) also known as Sokoto Gudali and Bunaji (BJ) also known as White Fulani of similar ages in selected farms in Adamawa State were used for the experiment. The animals used were in their first and second parities and early lactation stage (1-60 days). Eighty (80) blood samples (3ml) were collected from experimental animals via their jugular vein using 5 ml syringe and needle into a 5ml EDTA vacutainer tube and stored in an ice pack. Each tube was gently mixed by inversion, labeled with breed number, and transferred to the laboratory for DNA extraction at African Bioscience Laboratory (Ibadan, Nigeria).
DNA extraction was carried out using ZR-96 Genomic DNA miniprep®. Nanodrop 1000® spectrophotometer was used for the determination of DNA quality and quantity using a Roche DNA purification kit (Roche Diagnostics GmbH, Mannheim). Cell lysis, DNA binding, DNA washing and elution was carried out according to the manufacturer’s instructions.13
Cycling conditions was as follows: initial denaturation at 94oC for 4 minutes followed by 30 cycles 94oC for 30 seconds, 56oC for 30 seconds, 72oC for 2 minutes and a final volume contained 25ng genomic DNA, 1µM primers, 2mM MgCl2, 1U Platinum® Taq polymerase, and 1 x PCR buffer (Invitrogen Life Sciences, Dublin, Ireland). PCR products be purified and sequenced.
The sequence of primers14 were used for amplification of exons of HSP90AA1 gene in the present study is given in Table 1. The primers were obtained from Amnion Biosciences Pvt Ltd, Bangalore and reconstituted with distilled water to make stock solution of 100 pmol/μl and stored at -20 ºC. Ten micro litre of the stock solution was taken and diluted with 90 μl distilled water to obtain the working concentration 10 pmol μl.
|
Primer code |
Sequence (5’-3’) |
No. of base |
Annealing temperature (0C) |
Melting temperature (0C) |
Amplicon size (bp) |
|
KX-592814.1 |
F- GCGTCATCACGTGTCATCTT |
20 |
54 |
58.65 |
450 bp |
|
R- CCTCCTTTGGGGTTCCAGT |
19 |
58.84 |
Table 1 Primer Sequence, targeted region and amplicon sizes of Bovine HSP90AA1 gene in experimental cows
Source: (Kumar et al., 2015)
Agarose gel electrophoresis was carried out for testing the template DNA and PCR product. For checking the template DNA, 0.7 percent agarose gel was used (15). The gel was then visualized under UV trans-illuminator and photographed using gel documentation system (Figures 1 & 2). The DNA fragment sizes were determined in comparison to DNA molecular weight marker (1000 bp ladder) using Image Lab version 4.0 software.15
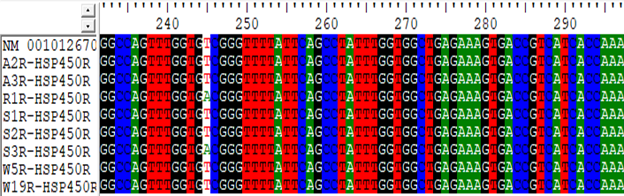
Figure 2 Sample of single nucleotide polymorphisms identified using NM_001012670.1 as reference HSP90AA1 gene.
A 965 bp fragment of exons of HSP90AA1 gene were amplified by Polymerase Chain Reaction. The sequence of primers14 were used for amplification of exons of HSP90AA1 gene. The primers were obtained from Amnion Biosciences Pvt Ltd, Bangalore and reconstituted with distilled water to make stock solution of 100 pmol / μl and stored at -20 ºC. Ten micro litre (10 μl) of the stock solution was taken and diluted with 90 μl distilled water to obtain the working concentration 10 pmol /μl.
Sequencing of HSP90AA1 was done and submitted to the gene bank. A reference sequence was downloaded from Ensembl while the HSP90AA1sequences were cleaned and edited using Bioedit® software. The HSP90AA1 sequences were aligned with reference sequence to identity the SNPs using the clustal X hosted in Bioedit® software. Genetic diversity indices were estimated using DNAsp software while allelic and genotypic frequencies were estimated by code written in R software.16
Variant prediction effect on the SNPs were carried out using Ensembl©. Stepwise discriminant procedure (STEPDISC) was also applied to determine, which performance traits were used in the final clustering analysis, this procedure determined the variables that had more discriminating power than the others. These distances were used to construct a dendrogram using the un-weighted pair group method analysis implemented in R 2.13.0.16
Table 2 shows the genetic diversity of the studied population. In this study, four Nigerian indigenous breeds of cattle were sequenced (Adamawa Gudali, Rahaji, Bokoloji and Bunaji) and the sequence generated from the four breeds were deposited in the GenBank with accession numbers MZ355888–MZ355895. There was a total of six (6) polymorphic informative sites scored in the sequences that defined four haplotypes. Five of the identified informative sites were found in Adamawa Gudali while one was found in Bokoloji. The total number of nucleotide difference and nucleotide diversity were six with Adamawa Gudali recording the highest (five out of six) and (0.014) respectively.
|
Breed |
N |
TM |
PIS |
Hd |
N |
K |
|
Bokoloji |
3.00 |
1.00 |
1.00 |
0.67 |
0.00 |
0.67 |
|
Adamawa Gudali |
2.00 |
5.00 |
5.00 |
1.00 |
0.01 |
5.00 |
|
Rahaji |
1.00 |
0.00 |
0.00 |
0.00 |
0.00 |
0.00 |
|
Bunaji |
2.00 |
0.00 |
0.00 |
0.00 |
0.00 |
0.00 |
|
Overall |
8.00 |
6.00 |
6.00 |
1.67 |
0.01 |
5.67 |
Table 2 Genetic diversity indices for four breeds of Nigerian indigenous cattle
N, Number of sequences; TM, Total number of mutations; PIS, Polymorphic informative sites; Hd, Haplotype (gene) diversity; N, nucleotide diversity; k, Average number of nucleotide differences.
The nucleotide and haplotype diversities, are the two most often used indices in assessing the level of genetic diversity of populations or of a gene in a population.17,18
The high haplotype diversity observed in Adamawa Gudali implied that the population of Adamawa Gudali have the greatest genetic diversity for HSP90AA1 gene which could be an advantage in a heat stress conditions.15 A strong haplotype diversity both within and between breeds also arise from multiple sites having shared ancestry, evolutionary adaptation, and diverse environmental disease exposures.18,19,20
The high value of polymorphic sites (mutations) in Adamawa Gudali is an indication of its capacity to generate enough adaptable ability to endure different stresses due to environment, management systems, heat and cold stress as compared to the other studied indigenous breeds.21
The lowest haplotype and nucleotide diversities, average number of nucleotide difference observed in Nigerian cattle were in Rahaji (0.00). The overall haplotype diversity observed in the Nigerian cattle was 0.8095. The low haplotype and nucleotide diversities recorded in this study in Rahaji and Bunaji (0.00) could be an indication of gene flow due to hybridization between populations 18,22 and could be due to the few numbers of samples sequenced in these studies since animals that are leading in milk yield and raised in different environments tend to have high nucleotide diversity.18,22
The gene flow estimate between the studied breeds of Nigerian indigenous cattle is presented in Table 3. Three breeds were used (Rahaji was excluded because only one sequence was obtained for Rahaji and one sequence cannot be used to generate Fst). There was a significant gene flow estimate between Adamawa Gudali and Bokoloji as well as between Bokoloji and White Fulani populations (0.67) except for Adamawa Gudali and Bunaji (0.00). For the genetic diversity distribution between populations (Gst), the highest distribution was between Bokoloji and Bunaji (0.447). The number of net nucleotide substitution per site between populations (Da) ranged from 0.000 between Adamawa Gudali and Bokoloji to 0.02446 between the other populations.
|
Population 1 |
Population 2 |
Hs |
Ks |
Kxy |
Gst |
Delta St |
Gamma St |
Nst |
Fst |
Dxy |
Da |
|
Adamawa_Gudali |
Bokoloji |
0.67 |
2.4 |
2.83 |
-0.1 |
0 |
0.34 |
0 |
0 |
0.01 |
0 |
|
Adamawa_Gudali |
Bunaji |
0 |
2.5 |
11.5 |
0.33 |
0.01 |
0.8 |
0.79 |
0.78 |
0.03 |
0.03 |
|
Bokoloji |
Bunaji |
0.67 |
0.4 |
9.33 |
0.45 |
0.01 |
0.94 |
0.97 |
0.96 |
0.03 |
0.03 |
Table 3 Gene flow estimates between breeds excluding Rahaji due to number of sequences available in Rahaji breed
Fst is a measure of genetic differentiation among population23 and it’s mostly represented as an inverse of Gst. They explain the level of polymorphism among biallelic markers. The zero (0.00) value of Fst recorded between Adamawa Gudali and Bokoloji in this study implies that there is low genetic differentiation between the two populations.24,25
This is in agreement with the findings of 17,25 who reported a low genetic differentiation between breeds of cattle within a continent. The low genetic differentiation observed in the Nigerian Breeds studied could be due to selection of those breeds for specific type of production which shaped the genome of the cattle breeds used in this study and the fact that these breeds are not widely spread as other continental breeds.18
In most cases, one hardly finds the mixture of Adamawa Gudali and Bokoloji as they both move in separate colonies. The high Fst values reported in this study for Bunaji with Adamawa Gudali and Boloji (0.78261 and 0.96429) respectively, suggest that only 4 to 22% of all variability is due inbreed diversity and 78 to 96% is due to interbreeding diversity.23,26
It also suggests that there is high genetic differentiation (the measure of allele frequency in HSP90AA1 between the population was high) between the population since Fst value ranges from 0 – 1 (22, 23, 24). The valued of Fst obtained in this study is higher than the ones reported in earlier studies (22) where they reported Fst values of 0.253, 0.152 and 0.143 for cattle, pigs, and goats, where intra-breed diversity was 74.7%, 84.8% and 85.75, respectively.23
The average number of nucleotide substitution per site between populations (Dxy) in this study ranged from 0.0077 between Adamawa Gudali and Bokoloji and 0.03125 between Adamawa Gudali and Bunaji. The number of net nucleotide substitution per site recorded in this study between populations (Da) ranged from 0.000 between Adamawa Gudali and Bokoloji to 0.02446 between the other populations. Number of mutational steps between haplotypes (Nst) ranged from 0.00108 to 0.96483and average number of differences between two population (Kxy) ranged from 2.83333to 11.5. The high gene flow recorded in this study agree with the report of 27 who reported high gene flow between the cattle populations in Cameroon and Nigeria. The high gene flow between the studied breeds may lead to a loss of genetic diversity through uniformity and a reduction of opportunities for future breed development.26 The genetic diversity levels if African Zebus have been reported to be high.28,29
This could be due to proximity of the population, purpose of breed development, the management and production systems in use which is mainly pastoral and traditional which support uncontrolled flow of genes 27,30 If the high gene flow continues without check, it will result to loss of the distinction we have among Nigerian breeds of cattle which will also bring about loss in global breed diversity.27 To reduce this high gene flow within the Nigerian breeds, there is need to maintain practical pastoral system management to limit uncontrolled genetic exchanges between breeds and increase productivity.27
Table 4 present the single nucleotide polymorphism (SNP) on HSP90AA1 protein in four breeds of indigenous cattle. The sequences generated in this study revealed six (6) polymorphic sites in the coding regions (136 G>A, 136 G>A, 89 C>G, 89 C>G, 86 A>G and 86 A>G). The amino acid substitution in this study were obtained from alignment with the wild type alleles of the putative coding regions. Of all the six, all were missense variants (A genetic alteration in which a single base pair substitution alters the genetic code in a way that produces an amino acid that is different from the usual amino acid at that position) with moderate impacts. Two in the mutations observed were deleterious in nature (21: 66943881; G> A) both at position 406 with frequency of 0 and 0.02.
|
Location |
Allele |
Consequence |
Impact |
Biotype |
CDS Position |
Protein Position |
Amino acids |
Codons |
SIFT |
SIG |
|
21_66943881_G/A |
A |
missense |
Moderate |
protein_coding |
406 |
136 |
L/F |
Ctc/Ttc |
deleterious (0.00) |
- |
|
21_66943881_G/A |
A |
missense |
Moderate |
protein_coding |
406 |
136 |
L/F |
Ctc/Ttc |
deleterious (0.02) |
- |
|
21_66944022_C/G |
G |
missense |
Moderate |
protein_coding |
265 |
89 |
V/L |
Gtc/Ctc |
- |
- |
|
21_66944022_C/G |
G |
missense |
Moderate |
protein_coding |
265 |
89 |
V/L |
Gtc/Ctc |
- |
- |
|
21_66944030_A/G |
G |
missense |
Moderate |
protein_coding |
257 |
86 |
V/A |
gTt/gCt |
tolerated (0.38) |
- |
|
21_66944030_A/G |
G |
missense |
Moderate |
protein_coding |
257 |
86 |
V/A |
gTt/gCt |
tolerated (0.36) |
- |
Table 4 Single nucleotide polymorphisms prediction effects on the HSP90AA1 protein
The change in the amino acid G>A on 21: 66943881 suggest that the SNPs will affect the normal function of the proteins at those positions. HSP90AA1 as one of the molecular chaperons of HSP gene sub-family play an important role in animals’ survivability, particularly in thermoregulation in conditions where heat stress is a challenge.31
Out of the six amino acids sequences, two were tolerated meaning that the substitution of such amino acids would not impair protein function while the two that were deleterious indicates that the substitution of such amino acids would in turn bring about harmful effect on the normal function of the amino acids.32
This type of mutation can bring about a complete translation of the original protein and this may cause a stop codon to be read which truncates further synthesis of protein and result in abnormal biological activities.33,34 Thermo-tolerance has been identified a quantitative trait that is influenced by genomic regions at the target gene that is important for thermoregulation through genomic studies in cattle35,36.
Chaudhari37 also reported distribution of SNPs within targeted regions of Bovine HSPB1 gene and reported a complete translation of thymine to cytosine at location 208395876 and silent transversion of guanine to thymine at location 723061520 within the coding regions in Karan-Fries and Sahiwal cattle. SNPs located at position 660 in the flanking region of HSP90AA1 was found be to be associated with several thermal conditions in Ovine species.38
In a similar study on Nigerian indigenous breed of cattle31 four distinct SNPs in HSP90 gene where thymine changed to Guanine at 116 (T116G) in Red Bororo, Guanine changed to cytosine at (G220C) in Sokoto Gudali, Guanine changed to Adenine at (G346A) in Ambala (Adamawa Gudali) and White Fulani. In an earlier study also, two SNPs of HSPA1A90 (trans-version and transition) led to a change in amino acid from Glycine to Alanine (G2033C) that was responsible for change in milk traits in Chinese Holstein cattle. Single Nucleotide Polymorphism in HSP90 were reported to be detected in coding regions of Sahiwal, Doeni and Holstein-Friesian crossbred cattle.15,39
The partial coding sequence obtained in this study (CDS 257-406) concur with the finding of Yakubu40 who reported that sequence length can vary with location from where the sequences are found. Also, the variation in sequence length within species recorded in this study could be due to differences in the genomic region where the sequences were obtained from, differences in evolutionary origin and differences due to complete coding sequences (CDS) or partial CDS.40,41
Also, possibility abound where variation in sequence length may arise due changes within the chromosomes such as DNA duplication, arrangement, insertion, or deletion of sequences.41,42 These variants may be an important tool for improving animals in the face of climate change with the goals of efficient feed utilization, productive and reproductive performance.31,43
The analysis of molecular variance of the studied breeds is presented in Table 5. The highest percentage variation is among breeds (58.18%) while that of within breed is (41.82%). This implies that there is less genetic variation within the studied breeds for HSP90AA1 gene and that more genetic variation is found among the population than within the individuals within the breeds studied.36,44
|
Level of variation |
Sum of squares |
Mean squares |
Percentage variation |
|
Among population |
17.87 |
2.26 |
58.18 |
|
Within population |
6.5 |
1.63 |
41.82 |
|
Total |
24.38 |
3.89 |
100 |
Table 5 Analysis of molecular analysis for four breeds of Nigerian cattle
AMOVA can be used to detect population clustering in a genetic dataset.45,46 Population genetics (Hardy-Weinberg law) postulates that a population will maintain the same allelic frequency.24 It can be used to determine genetic differentiation among populations.44
The high percentage of variation recorded among populations gives an indication of the variation present among a population of the studied population for the HSP90AA1 gene. The low rate of the within population (41.818%) recorded in this study concur with the findings of Chaudhari37 who reported lesser genetic diversity in the genomic region of Bovine HSPB1 in Bos indicus as compared to cross bred cattle. Domestic animal diversity is crucial for species adaptation in a climate change era, which comes with many difficulties in terms of specie adaptation and disease resistance.47
Contrary to the finding of this study, portioning of genetic variation was reported to be found among individuals within the breed rather than among the population.18 They reported that only 5.99 percent of the genetic variation was found among population while 94.01 percent of the genetic variation was found within a population.20
Also, in the study in goats, the percentage genetic variation was reported within the population than among the population.48 They reported that when populations were structured according to historical data, the AMOVA confirmed that most (87.34%) of the total variation was caused by differences between individual animals within populations.
Özşensoy and Kurar,44 also reported that a total genetic variation of 98.04% was found within populations, whereas 1.96% variation was recorded among the populations. Contrary to other findings, total genetic variations were obtained as 87% among populations.49 This agrees with the current studies. The genetic variation in HSP90AA1 recorded in this study can be explored for genetic improvement of the studied population through selection.50
A total of eight sequences were generated from the selected breeds and have been deposited in the GenBank with accession numbers MZ2355888 – MZ2355895. The sequences generated in this study revealed six (6) polymorphic sites in the coding regions that defined four haplotypes. There is more genetic variation among Nigerian breeds of cattle than within the breeds for HSP90AA1 gene.
None.
We acknowledge the contribution of Late Professor I. I. Adedibu in initiation, supervision and final production of the main thesis where this paper was extracted from.
The authors declare that there are no conflicts of interest.

©2024 Abbaya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.