Journal of
eISSN: 2373-4396


The ultraviolet (UV) radiation resistance-associated gene (UVRAG) was initially identified for its capability to partially complement UV sensitivity in xeroderma pigmentosum (XP) cells. Subsequently, UVRAG is recognized as a tumor suppressor and autophagy-related protein. In recent years, substantial bodies of work provide new insight into UVRAG structure and multiple biological functions. UVRAG consists of a proline-rich domain, a calcium-dependent phospholipid binding C2 domain, a coiled-coil domain, a DNA-dependent protein kinase binding domain and a CEP-63 binding domain. In addition to its tumor suppression activity, UVRAG regulates autophagy and endocytic trafficking, participates in apoptosis, maintains genomic stability, gets involved in organ rotation during development. In the heart, UVRAG is required to maintain cardiac structure and function. UVRAG deficiency in the heart leads to age-related dilated cardiomyopathy. In this review, we summarize these recent findings of UVRAG that are shedding new light on its crucial roles in many biological processes and implications for the pathophysiology of various diseases including heart disease.
Keywords: UVRAG, autophagy, endocytosis, epoptosis, cardiomyopathy, cardiac function
The ultraviolet (UV) radiation resistance-associated gene (UVRAG) was initially identified for its capability to partially complement UV sensitivity in xeroderma pigmentosum (XP) cells.1 The human UVRAG gene was localized to chromosome 11q13, a region implicated in human cancers such as colon cancer, breast cancer and gastric cancer.1 Consistently, UVRAG is frequently monoallelically deleted or mutated in these cancers, indicating a tumor suppressor activity.2–4 UVRAG has been shown to suppress tumor growth by macroautophagy (hereafter refer to as autophagy), a degradative pathway for the bulk degradation of cytoplasmic components such as misfolded proteins, protein aggregates and damaged organelle.5 In addition to tumor suppression function, further characterization of UVRAG in recent years provides new insight into its structure and crucial roles in the regulation of multiple cellular processes. Here, we summarize these recent findings of UVRAG, with an emphasis on its physiological function in the heart.
UVRAG structure
UVRAG contains a proline-rich (PR) domain, a calcium-dependent lipid-binding C2 domain, a coiled-coil domain (CCD) and a C-terminal domain (Figure 1).6,7 The PR domain mediates the interaction of UVRAG with phosphorylated Bif, which promotes autophagosome formation.8,9 The C2 domain binds phospholipid and Vps16, a component of class C Vps complex. The C2 domain also binds Bax.10 The CCD domain allows UVRAG to associate with Beclin 1, which induces Vps34 activity and regulates autophagosome formation, autophagosome maturation and endocytic trafficking.5 The C-terminal region has previously been considered unstructured. However, a CEP-63 binding domain (CEP63-BD) and a DNA-dependent protein kinase binding domain (DNAPK-BD), which binds CEP-63 and DNA-PK, respectively, has recently been identified. The region harboring CEP63-BD (amino acids 269-442) cooperates with the C2 domain to bind Vps16.11
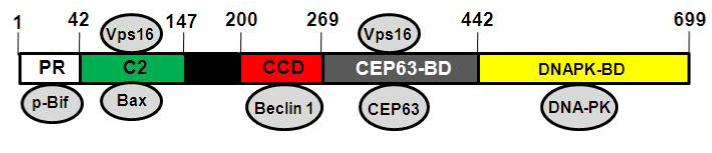
Figure 1 Schematic representation of domains of human UVRAG. The proline-rich (PR) domain binds phosphorylated Bif. The C2 domain associates with Bax. The Coiled-coil domain (CCD) interacts with Beclin 1, the CEP63 binding domain (CEP63-BD) with CEP63, the DNAPK binding domain with DNA-dependent protein kinase (DNA-PK). The C2 domain and the region harboring CEP63-BD (amino acids 269-442) cooperate to bind Vps16.
UVRAG regulates autophagy and endocytic trafficking
In mammalian cell, Vps34, Beclin 1 and p150 form a core complex and function in membrane trafficking such as autophagy and endocytosis.12,13 UVRAG binds Beclin 1 through its CCD domain and up-regulates Vps34 activity, which leads to increase of phosphatidylinositol 3-phosphate (PI3P) and enhanced autophagosome formation.5 CDK5-mediated phosphorylation of Bif (also known as endophilin B1 or Bax-interacting factor 1), a UVRAG interacting protein, is required for activating Vps34 and inducing autophagosome formation.14 The direct interaction of PR domain of UVRAG and SH3 domain of Bif is responsible for the interaction of these two proteins.8 It has been suggested that UVRAG binds to Vps34 core complex and promotes autophagosome and endosome maturation, which is inhibited by Rubicon.15 UVRAG has been demonstrated to bind Vps16, a component of C-Vps/HOPS complex, to recruit the membrane fusion machinery to promote autophagosome maturation and later step of endocytosis (Figure 2). This function of UVRAG is independent of Beclin 1.7 Recently two new studies suggest that, UVRAG does not bind to the STX17–HOPS complex, which is essential for autophagosome-lysosome fusion, suggesting that UVRAG may not be directly involved in autophagosome maturation.16,17
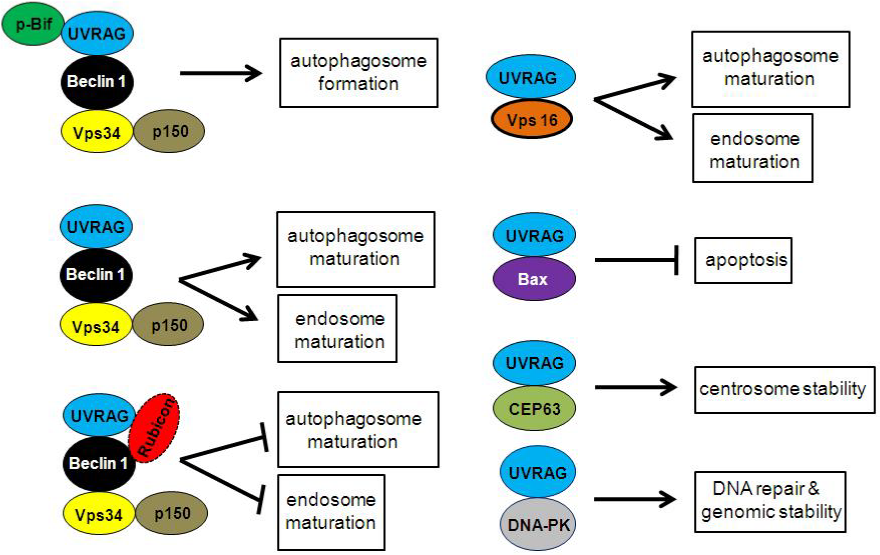
Figure 2 UVRAG complexes. UVRAG directly binds phosphorylated Bif, Beclin 1, Bax, CEP63 and DNA-PK, forming distinct protein complexes to regulate autophagosome formation, autophagosome maturation, endosome maturation, apoptosis, centrosome stability, DNA repair and genomic stability.
UVRAG participates in apoptosis
Given the crosstalk between autophagy and apoptosis, it is not surprising that UVRAG is involved in apoptosis. However, Yin et al.,10 find a novel mechanism by which UVRAG induces apoptosis independent of autophagy. UVRAG binds BCL2-associated X protein (BAX) through C2 domain and prevents Bax from translocating to membrane of mitochondria, which inhibits the reduction of mitochondria membrane potential, cytochrome C release and activation of caspase-9 and caspase-3 (Figure 2). Therefore, UVRAG is essential in the intrinsic pathway for apoptosis by sequestering Bax from inducing apoptosis.10 UVRAG-mediated suppression of apoptosis is implicated in cancer therapy. In cancer cells, chemotherapy and radiation up-regulate UVRAG. Cancer cells deficient for UVRAG are more susceptible to apoptosis induced by chemotherapy and radiation.10 Further understanding of UVRAG and Bax interaction may lead to therapeutic targets against human cancers.
UVRAG maintains chromosome stability
Autophagy is required to sustain genomic integrity under metabolic stress. Compromised autophagy fails to maintain metabolism, which is associated with increased DNA damage, gene amplification, and aneuploidy.18,19 Zhao et al report that UVRAG preserves genomic stability and centrosome integrity. Surprisingly, these capabilities of UVRAG are independent of its function in autophagy. Instead, a portion of UVRAG is recruited to centrosome and sustains centrosome integrity by binding CEP-63, a component of centrosomes, through CEP63-BD localized to C-terminal domain. In addition, UVRAG binds and activates DNA-PK through DNAPK-BD located in the C-terminal domain inside the nucleus and promotes DNA double-strand-break repair (Figure 2). As centrosome aberrations and chromosomal instability are hallmarks of human cancer, UVRAG may confer tumor suppressor activity independent of autophagy.11
UVRAG and organ rotation
In Drosophila, UVRAG is reported to be involved in the control of organ rotation, a process essential for the establishment of left-right body asymmetry during development. Drosophila mutant deficient for UVRAG shows failure of normal organ rotation, impaired endocytosis, but normal autophagy. Notch signaling is enhanced by attenuated endocytic degradation caused by UVRAG deficiency. UVRAG mutants with compromised Notch signaling rescue the rotation defects.20 Interestingly, a heterotaxy patient with complex cyanotic heart defects, altered lung lobation, symmetric liver, and abnormally lobulated spleen (polysplenia) exhibits a inv (11) (q13.5;q25) inversion. UVRAG, maps to 11q13, is disrupted by the chromosomal rearrangement in this patient.21 In addition, the Notch 1 protein in the cells isolated from the UVRAG-mutated heterotaxy patient is more abundant than normal patient.20 Overall, these data suggest a role of UVRAG in organ rotation.
Essential role of UVRAG in the heart
Recently, we explore the role of UVRAG in the heart using a mouse model deficient for UVRAG generated by piggyBac (PB) transposition.22,23 The mouse UVRAG gene is located on chromosome 7 and contains 15 exons interrupted by 14 introns. The PB transposon is inserted into intron 14 and the disruption of UVRAG gene expression is confirmed at both the mRNA and protein levels. UVRAG-deficient mice are viable, fertile and exhibit normal cardiac morphology and function at 2 and 6 months of age. However, at 10 months of age, the hearts from UVRAG-deficient mice are larger than the corresponding normal controls (Figure 3A). Moreover, UVRAG deficiency in the heart leads to interstitial collagen accumulation (Figure 3B), increased cross-sectional area of individual cardiomyocytes, elevated expression of cardiac fetal genes and enhanced apoptosis. In addition, pro-inflammatory cytokine expression is increased in the hearts from UVRAG-deficient mice. Echocardiography reveals that UVRAG-deficient mice at 10 months of age have significantly greater left ventricular end systolic diameter (LVESD) and left ventricular end diastolic diameter (LVEDD) (Figure 3C). Furthermore, fractional shortening and ejection fraction, two indices of cardiac function, are attenuated in 10-month-old UVRAG-deficient mice. These results demonstrate that UVRAG-deficient mice develop age-related dilated cardiomyopathy with compromised cardiac function. Further characterization reveals that autophagosome is accumulated and p62 protein levels are elevated in UVRAG-deficient heart. Assessment of autophagic flux suggests that autophagy process is impaired in the hearts from UVRAG-deficient mice. In agreement with these observations, autophagosomes are accumulated in UVRAG-deficient MEFs, which is caused by impaired autophagic flux. Overall, the data in this study suggest that UVRAG is essential in autophagic flux and the maintenance of cardiac structure and function.23
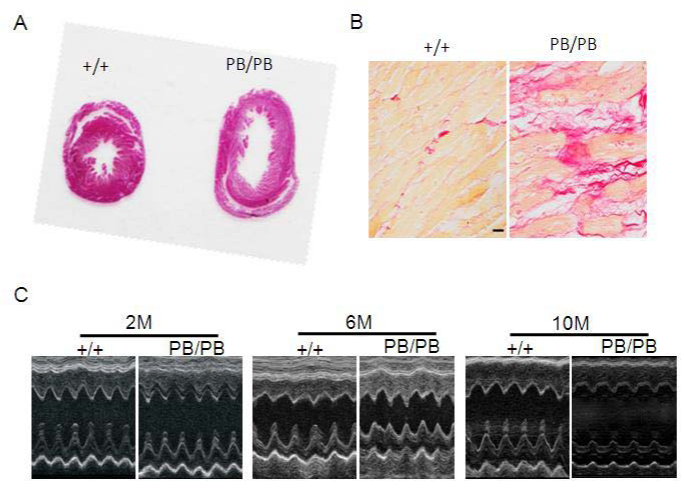
Figure 3 UVRAG-deficient mice develop age-related cardiomyopathy associated with compromised cardiac function.
(A) Cross-sectional view of the hearts from 10-month-old WT and UVRAG-deficient mice.
(B) Picrosirius red staining of paraffin sections of the hearts from 10-month-old WT and UVRAG-deficient mice. Scale bar: 40 mm.
(C) Representative M-mode echocardiographs from 2-, 6-, and 10-month-old UVRAG-deficient mice and the corresponding WT controls. +/+: WT mice; PB/PB: UVRAG-deficient mice.23
This study poses a few interesting questions. What is the role of UVRAG in autophagosome formation in the heart? Is endocytic trafficking dysregulated by UVRAG deficiency in the heart and contributes to compromised cardiac function? Is apoptosis caused primarily by UVRAG deficiency or secondary to impaired autophagy? What are the precise mechanisms by which UVRAG deficiency up-regulates pro-inflammatory cytokines in the development of age-related cardiomyopathy? Does UVRAG deficiency in non-cardiac organs have impact on cardiac function? Does UVRAG deficiency have effect on myocardial capillary or coronary morphology or density? How is UVRAG regulated and what role does it play under various cardiac pathological conditions such as pressure overload, ischemia and reperfusion? More extensive work is required to elucidate these questions in the future to better understand the physiological and pathophysiological function of UVRAG in the heart and related diseases, which may become an integral part of drug discovery.
UVRAG forms complexes by binding different partners. These complexes exert various biological functions such as regulation of autophagosome formation, autophagosome maturation, endocytic trafficking, apoptosis, centrosome stability, DNA repair and genomic stability. By using UVRAG-deficient mice generated by piggyBac transposition, we find that UVRAG is required for autophagy in the heart and the maintenance of cardiac function. Given that UVRAG plays crucial roles in multiple cellular processes, it could be served as a potential therapeutic target for a variety of diseases including certain types of heart disease.
None.
Authors declare that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.