Journal of
eISSN: 2572-8466


Research Article Volume 3 Issue 3
1Department of Biological Sciences, Madurai Kamaraj University, India
2National Research Centre for Banana, India
3Department of Plant Biotechnology, Madurai Kamaraj University, India
Correspondence: Jebasingh T, School of Biological Sciences, Madurai Kamaraj University, Madurai-625021, Tamil Nadu, India, Tel 91-9043568671
Received: February 14, 2017 | Published: June 12, 2017
Citation: Jebasingh T, Backiyarani S, Manohari C, et al. Transformation of cardamom with the RNA dependent RNA polymerase gene of cardamom mosaic virus . J Appl Biotechnol Bioeng. 2017;3(3):326-329. DOI: 10.15406/jabb.2017.03.00066
Cardamom (Elettaria cardamomum Maton) is an important spice crop. It is affected by Cardamom mosaic virus (CdMV). In order to make cardamom plants resistant to CdMV by the pathogen-derived resistance approach, the RNA dependent RNA polymerase gene (NIb) of CdMV in the plant expression vector pAHC17, was introduced into cardamom embryogenic calli along with GFP-BAR by particle bombardment. Transformants were selected on a medium containing bialaphos and the presence of NIb and gfp genes in cardamom plants were confirmed by PCR, Southern hybridization and GFP expression.
Keywords: cardamom, particle bombardment, transgenic, GFP-BAR
CdMV, cardamom mosaic virus; PVY, potato virus Y; PSbMV, pea seed-borne mosaic virus; WYMV, wheat yellow mosaic virus; Nib, nuclear inclusion protein-b; GFP, green fluorescent protein; PCR, polymerase chain reaction; BAP, 6-benzyl amino purine NAA, naphthalene-3-acetic acid; PPV, plum pox virus
Small cardamom (Elettaria cardamomum Maton.) is considered as the third most extensive spice in the world after saffron and vanilla. It is popularly known as the queen of spices. It is one of the major exporting spice crops of India. The production of cardamom has been greatly affected by a few viral diseases. Cardamom mosaic disease, also known as katte (meaning disorder) is the most widespread in south India, caused by Cardamom mosaic virus (CdMV). It is classified as a new species of the Macluravirus genus of family Potyviridae.1 CdMV has already affected almost 75 % of the cardamom cultivation in India. The impact of the disease reached its peak in 1987 when the export was reduced to 12 % of that of the previous year.2 This resulted in a loss of Rs. 60 millions in foreign exchange. There is a noticeable reduction in the area of cardamom cultivation especially in Karnataka and Tamil Nadu in the recent years. The disease has become widespread, as there is no effective means to control the transmission of the virus. Moreover, management of viral diseases through chemicals is not possible in cardamom and till now none of the cardamom cultivars has been identified as resistant to CdMV. Therefore, it becomes imperative to develop CdMV-resistant plants through genetic transformation.
Nuclear Inclusion b (NIb) gene of potyviruses encodes an RNA dependent RNA polymerase (RdRp) for the replication and production of progeny viral RNA. Few reports are already available for the virus resistance with the NIb gene of Potyviridae. Reports of replicase-mediated resistance to Potato virus Y (PVY)3 concluded that the resistance mechanism was protein-based rather than RNA-based because the resistance phenotype was influenced by mutations affecting the primary structure of the protein encoded by the transgene. Nicotiana benthamiana plants carrying intact and mutated NIb of Plum pox virus (PPV) showed some degree of protection when low inoculum was given. The delayed, very specific, highly resistant phenotype was observed in plants carrying a Gly to Val mutation at the GDD motif.4 The protection was mainly mediated by RNA through an “induced” co-suppression mechanism. Virus-induced resistance in transgenic pea carrying Pea seed-borne mosaic virus (PSBMV) replicase sequence was due to gene silencing.5 Transgenic wheat with the NIb gene of Wheat yellow mosaic virus (WYMV) has shown resistance against the WYMV was not due to NIb derived siRNA but due to disruption of a host gene required for WYMV infection.6 Similar resistance mechanism is observed for PVY in the transgenic potato with truncated NIb gene.7 Here we report the bombardment of NIb gene of CdMV along with marker genes (Green Fluorescent Protein (GFP) and BAR (bialaphos resistance encodes for Phosphinothricin Acetyl Transferase that detoxifies phosphinothricin (PPT)) into embryogenic calli derived from the high yielding “Green gold” variety of cardamom. The plants regenerated from the bombarded calli were confirmed for the presence of NIb and the reporter gene gfp by PCR.
Cloning of NIb gene in pAHC17 vector
The 1.7 kb NIb coding region of CdMV with ATG was released from pNIb18 with Bam HI digestion and cloned at the corresponding site of the plant expression vector pAHC179 to derive pAHC-NIb. The correct orientation of the insert was confirmed with Hind III digestion.
Plant material
Rhizomes of green gold variety were used as explants for callus initiation. The surface sterilization of rhizome, callus initiation and embryogenic calli formation were done according to Manohari et al.10 All media were based on MS basal medium11 with pH adjusted to 5.8 before autoclaving.
Gene transfer
The gene transfer is done with Biolistic PDS-1000/He system uses high-pressure helium, released by a rupture disk, and partial vacuum to propel a macro carrier sheet loaded with millions of microscopic tungsten or gold micro carriers toward target cells at high velocity. The micro carriers are coated with DNA used for transformation. The macro carrier is halted after a short distance by a stopping screen. The DNA-coated micro carriers continue traveling toward the target to penetrate and transform the cells. Micro carrier preparation was performed according to Sanford et al.12 6 mg of gold particles (1μm diameter) were coated with pAHC-NIb (5μg) and pGFP-BAR (5μg).13 The embryogenic cardamom calli (30-40 numbers) to be bombarded were arranged in a circle of 2 mm diameter on the center of a petri dish containing osmotic medium (MS with 8.8μMBAP (6-benzyl amino purine), 0.5μMNAA (Naphthalene-3-acetic acid), 0.25M Mannitol, 0.25M Sorbitol in 0.25 % phytagel). The calli were incubated in dark for 4 hrs prior to bombardment. Three bombardments were done with PDS-1000/He Biolistic® Gene Delivery system (Bio-Rad, Hercules, CA). Rupture disk (1100 psi) was mounted on the gas acceleration tube. DNA-coated gold particles were applied onto the center of the macro carrier disk that was mounted on the macro carrier holder and allowed to dry. A petri dish with cardamom embryogenic callus was kept on the petri dish holder at a distance of 9 cm. The chamber pressure was maintained at 27 inches of Hg (9.5 Kpa absolute pressures).
Cardamom regeneration and establishment of regenerated plants
After bombardment, the embryogenic calli (107 numbers) were incubated in dark at 25 ± 2°C for 24 hrs and then transferred to selection medium, containing bialaphos (4 mg/l). The cultures were sub-cultured on fresh selection medium at regular intervals of 4 weeks and incubated in the dark. After two sub-culturing, the bombarded calli were transferred to regeneration medium (MS medium with 4.4μM BAP and 0.5μM NAA). The calli were incubated in 16hrs light (50μE m-2 sec-1) (provided with cool white fluorescent tubes)/ 8-hours dark condition. The temperature in the culture storage room was maintained at 25 ± 2°C. After transferring the calli to fresh medium every month, the calli started showing signs of shoot formation in about 3 months time. At this stage, the calli were transferred to MS media with concentrations of 13.2μM BAP and 0.5μM NAA to develop shoots and roots. Fully developed rooted plantlets were removed from the culture bottle, thoroughly rinsed with running water and transferred to cups containing a mixture of sterilized vermiculite and sand (1:1) and maintained in a plant growth chamber for two weeks under controlled temperature (23 °C) and 16/8h light/dark photoperiod and 90% relative humidity for hardening. The plants were initially irrigated with ¼ strength of MS medium on alternate days, followed by tap water after 1 week. Thereafter, the plants were transferred to a mist chamber in a greenhouse.
DNA extraction, Dot blot, PCR and Southern analysis
DNA was extracted from 86 putative transgenic cardamom plants maintaining in growth chamber by a modified Dellaporta DNA extraction method.14 Dot blot was performed for 86 plant DNA with 1.5 Kb NIb probe made from pNIb1 (Jebasingh et al., 2008) with NIb mitrev (5’-GACTCCAAAAATACCATTG-3’) and NIbEF1 primers (5’-GTTTACGAGGATCCATGTCAA-3’) (Figure 1). PCR was done for 86 putative transgenic plants with NIbEF1 and NIb mitrev primers (Figure 1). The reaction mixture contained 2μl of 10× PCR buffer, 1 U Taq DNA polymerase (MBI, Fermentas), 2 pmol of each primer, 2mM dNTPs in a final volume of 20μl. The thermal profile of the reaction was: initial denaturation at 94°C for 2 min, 35 cycles at 92°C for 30 s, 48°C for 30 s, 72°C for 30 s, and finally 72°C for 8mins. The gfp gene was amplified as per the above procedure except for the primers and the annealing temperature. The 0.8 kb gfp was amplified with the primers GFP. F(5’-AGGCCTCATGGTGAGCAAGGGCGAGG-3’) and GFP. R (5’- CCATGGCCGCTTTAC TTGTACAGCTCGTC-3’) at an annealing temperature of 50°C. After PCR, the amplified products were separated on 1.5% agarose gels and transferred to Hybond N+ nylon membrane and Southern analysis was done for both NIb and gfp with the corresponding probes.
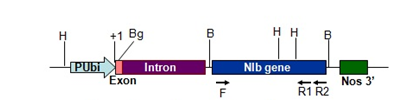
Figure 1 Linear map of pAHC-NIb. PU bi-Ubiquitin promoter, F-NIbEF1 primer, R1-NIbER1 primer, R2-NIbmutrev primer, H-HindIII, Bg-BglII, B-BamHI, +1-Transcription initiation site.
Cloning of NIb from transgenic cardamom
Total high molecular weight genomic DNA was isolated from young NIb transgenic leaf tissue using Dellaporta DNA extraction method.14 The 1.5 kb NIb was amplified from plant number 104 (which was positive only for NIb gene), eluted and cloned in pXcmKn12.12 The clone was initially confirmed by Bam HI digestion and then the insert (1.5 kb) of the clone was sequenced with M13 reverse and forward primers
Visualization of GFP expression
GFP expression was observed under a florescence microscope for embryogenic calli (3 weeks post bombardment) and for leaf sections from transgenic plants. Leaves from non-transgenic plants were used as control.
Establishment of callus cultures
Callus induction was observed from the inner core region of the cardamom rhizome on MS medium supplemented with the plant growth regulators like 9.0 μM2,4-dichloro phenoxy acetic acid (2,4 D), 2.3 μM Kinetin and additives like 0.1% casein hydrolysate and 0.1% polyvinyl pyrrolidone (PVP) after a period of 30 days. The explants exhibited swelling within four weeks, followed by the development of whitish callus after another six to eight weeks. Large number of white, soft and friable calli were obtained after 60 days by sub-culturing twice in the same medium. After two subcultures, non-embryogenic, loose calli were obtained. These embryogenic calli were used for the bombardment.
Selection and multiplication of transformants
Non-transformed embryogenic cells were initially cultured on media containing 1 to 6 mg/l of bialaphos. Growth was significantly reduced at 4 mg/l concentration. Hence, after bombardment, without disturbance, the embryogenic cell masses were transferred to selection medium with bialaphos (4 mg/l). During selection on bialaphos-containing medium, untransformed calli gradually turned brown and bialaphos resistant calli remained embryogenic. After five weeks, the surviving embryogenic calli were viewed under a fluorescence microscope to rule out transient expression of GFP (Figure 2). The bombarded calli developed into plantlets after six months when they were sub-cultured in regeneration medium (Figure 3).

Figure 3
Analysis for NIb gene in putative stable transformants
DNA extracted from the 86 putative transgenic cardamom and non-transformed plants were used for dot blot analysis with the 3’ truncated NIb probe (1.5 Kb size) (Figure 1), as 0.2 kb at the 3’ region of NIb was shown to hybridize even to healthy plants.15 16 (plant numbers 1, 23, 35, 41, 71, 80, 91, 93, 97, 99, 100, 101, 103, 104, 106 and 107) out of the 86 plants were positive for 3’ truncated NIb probe in Southern hybridization (Figure 4). The 1.5 kb NIb amplicon from plant number 104 (NIb transgenic plant) was cloned and sequenced and the sequence showed 100% identity to the NIb gene of CdMV (Accession number: AJ345002).

Figure 4 Southern hybridization of transgenic plants with NIb probe (1.5kb fragment amplified from pAHC NIb with NIbEF1 and NIb mitrev primer (Figure 1)) NIb probe (lanes 1 & 23), non-transformed plants (lanes 13 & 22), transgenic plants 1(2), 2(3), 3(4), 23(5), 35(6), 41(7), 71(8), 80(9), 90(10), 91(11), 93(12), 97(14), 99(15), 100(16), 101(17), 103(18), 104(19), 106(20) & 107(21).
Analysis for gfp gene in putative stable transformants
The transgenic plants in the growth chamber were screened for gfp gene. Dot blot analysis was done with gfp probe. Plant numbers 23, 41, 85, 93, 97, 99, 103 and 106 were positive for gfp probe. Plant number 85 was positive only for gfp probe and not for NIb probe. The 0.8 kb amplicon from all these plants hybridized with the gfp probe in Southern blot (Figure 5, lane 5). The non-transformed plants were negative for gfp probe (Figure 5, lane 1). In order to observe the GFP expression, leaf sections were made from plant 85 (GFP transgenic plant) and non-transformed plant and observed under a fluorescence microscope (Figure 6).
The Nuclear inclusion b (NIb) gene of Potyviridae encodes RNA dependent RNA polymerase for the replication of the virus. The RNA dependent RNA polymerase has been used for potyvirus resistance in many crops.3-7 The production of cardamom in India has been greatly affected by Cardamom mosaic virus.2 “Green-gold” variety of cardamom has been reported to be a high-yielding variety.16 However this variety is highly susceptible to CdMV. Hence this variety was selected to develop pathogen-derived resistance against CdMV with the viral NIb gene. Transformation was done by particle bombardment in embryogenic calli with pAHC-NIb and pGFP-BAR. pGFP-BAR is a dual marker plasmid comprising the reporter gene gfp (green fluorescent protein) and the selectable bar gene (Basta tolerance).13 The disturbance of embryogenic calli sometimes could affect the regeneration of cardamom calli. Therefore after the bombardment, the bulk of embryogenic calli was taken and kept in the selection medium. Due to this, some embryogenic calli at the top could escape from the selection. A few transgenic plants were obtained with NIb alone without gfp. All the transformed plants raised from embryogenic calli were morphologically identical to non-transformed cardamom (Figure 3A & 3B). DNA from 86 transgenic cardamom plants was extracted and a dot blot analysis was done with 1.5 kb NIb probe as the full-length NIb probe (1.7kb) showed positive signal in the autoradiogram even with DNA from non-transformed plants due to the integration of a 0.2 kb fragment from the 3’ end of NIb gene in non-transformed cardamom and other plants.15
Plant number-85 which is not positive for NIb probe has only gfp gene. The cross section of this plant (gfp transgenic) and non-transformed plants showed GFP expression (Figure 6). The GFP expression in non-transformed plants could have been due to secondary metabolites in cardamom leaves since there was no GFP expression in the calli derived from non-transformed plants (Figure 2). Such a phenomenon of reduced amount of secondary metabolites was observed in the calli from Fabiana imbricata and Ephedra when compared to the whole plants.17,18 Therefore gfp is not the ideal reporter gene in the genetic engineering of cardamom plants. But it could be useful at the level of callus selection. Plants (35, 71, 80, 91,100, 101, 104 and 107) have NIb alone. Plants 23, 41, 93, 97, 99, 103 and 106 have NIb and gfp genes. In order to confirm the presence of NIb gene in the plant genome, 1.5 kb NIb was amplified from plant 104, cloned and sequenced. The sequence of 1.5 kb bases obtained showed 100% identity to the NIb gene of CdMV. Thus the stable transformants with NIb gene was obtained. The challenging of these NIb transgenic plants with CdMV to evaluate for virus resistance is in progress.
The NIb gene of Cardamom mosaic virus was cloned in plant expression vector and it was co-bombarded with pGFP-BAR into cardamom calli. GFP expression was observed in bombarded calli. The calli were regenerated into cardamom plants and were screened for the presence of gfp and NIb. Out of the 110 plants screened, one of them was found to contain only the NIb gene and two have both NIb and gfp. The NIb transgenic cardamom plants would be scored for resistance to CdMV.
The authors acknowledge the DBT and CSIR (Government of India), New Delhi for funding and fellowships. We are thankful to Dr. P Ravichandran for his helpful suggestions. Dr. Joseph Rajkumar is acknowledged for providing the cardamom rhizome.
The author declares no conflict of interest.

©2017 Jebasingh, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.