Journal of
eISSN: 2572-8466


Creative Review Volume 7 Issue 4
1Department of plant breeding and genetics, University of Agriculture, Pakistan
2Department of molecular biology, Virtual University, Pakistan
Correspondence: Iffat Nazir, Department of Plant Breeding and Genetics, Faculty of Agri Sciences, University of Agriculture Faisalabad, Pakistan
Received: June 05, 2020 | Published: July 23, 2020
Citation: Nazir I, Mahmood HZ. Polymerase chain reaction: a creative review. J Appl Biotechnol Bioeng 2020;7(4):157-159. DOI: 10.15406/jabb.2020.07.00228
In molecular biology, a scientific technique PCR (polymerase chain reaction) is used to generate thousands to millions of copies of a single particular DNA sequences to amplify a single or few copies of a piece of DNA across several orders of magnitude. For multiple applications, PCR is an ordinary and often vital practice used in medicinal and biological research labs and is used for diagnosis and investigation of multiple diseases. In PCR mainly three major steps are involved. Denaturation, annealing, and extension. PCR can be used to detect not only the human genome but also the genome of viruses and bacteria. PCR is especially useful in forensic laborites because a very small amount of original DNA is required. In the development of cancer, genes have been implicated through PCR.
Keywords: polymerase chain, human genome, annealing, extensión
PCR, polymerase chain reaction; DNA, deoxyribonucleic acid, Q-PCR, quantitative PCR
In molecular biology, a scientific technique PCR (polymerase chain reaction) is used to generate thousands to millions of copies of a single particular DNA sequences to amplify a single or few copy of a piece of DNA across several orders of magnitude. Polymerase Chain Reaction was firstly developed by American biochemist, Kary Mullis in 1984. Mullis received the Japan Prize and Nobel Prize in 1993 for developing Polymerase Chain Reaction. Gobind Khorana in 1971 describes the basic theory of DNA replication. For multiple applications, PCR is an ordinary and often vital practice used in medicinal and biological research labs and is used for diagnosis and investigation of multiple diseases. PCR has now become an indispensable technique used in biological and medical research labs for variety of applications because it is simple, quick, and inexpensive. PCR technique amplifies rapidly the specific DNA fragments from small quantities of source DNA material, even when that source DNA is comparatively poor quality. PCR is quick, easy, and fast method of amplification, it amplifies Millions of copies of any DNA fragment in a very short period
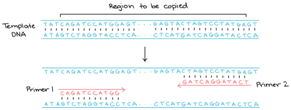
Basic concept of PCR
The basic principle about PCR is simple, quick, and easy. As the name indicates it is chain reaction. One molecule of DNA is used to produce 2 copies, then 2 molecules to produce 4 copies then 4 and 8 copies of DNA molecules and so on. The specific proteins called the Polymerases are responsible for continuous doubling of the DNA molecules. Polymerases are those enzymes that are responsible for long molecular strands by stringing the individual DNA building strand. For proper working of chain reaction polymerase requires DNA building blocks, a single nucleotide consists of four nitrogenous bases Adenine, Guanine, Thymine, Cytosine, and also need a small fragment of DNA known as a primer to which these building blocks are attached as a template to provide a new strand. PCR is a very efficient technique used to attain many copies of exact DNA, it means selectively amplifying a particular fragment of DNA. The fragment may characterize by small pieces of a large and complex piece of the genome for example a specific Exon of human genome. It is just like a molecular photocopier. PCR can amplify the usable amount of DNA in about 2 hours, the template DNA cannot be highly purified. PCR can amplify a single DNA molecule even from a single sperm. Polymerase Chain Reaction depends upon the ability of DNA – copping enzymes to stay stable at high temperatures. Mullis’s innovative PCR process is unproductive because it requires a lot of time, a huge amount of DNA polymerase, and constant attention throughout the PCR processes.1–5

Steps in PCR
Polymerase chain reaction mainly consists of three steps namely Denaturation, Annealing, and Extension. In the first step, Denaturation two strands of DNA denatured at a high temperature about (90-97 °C). In second step, primers anneal the template strand of DNA to further extension. At the last step it gets a multiple copies of the genome. Polymerase chain reaction is firstly heated at about 90- 97 °C temperature to denature the double-stranded DNA, next an enzyme called Taq polymerase synthesis is used to build two new DNA strands using the original strands as a template. This process results in Duplication processes in which one strand is a new one is old strand and so on… these new strands can further use to create new DNA copies and so on this process is repeated again and again to make millions of DNA copies. The annealing steps the second step in the PCR technique this step is very useful in forensic chemistry, this step happens at a lower temperature about 50- 60°C and this allows the primers to hybridize to their particular paired DNA strands.6–15 The newly synthesized DNA strands are then further used to make identical copies of the original DNA template strand. Taq polymerase used to add up the presented nucleotides to the end of the annealed primers. The extension of annealed primers occurs at approximately72°C for 2-3 minutes. The beauty of Polymerase chain reaction is that it is a very speedy method as compared to other techniques because each strand doubles the number of the copies of desired DNA strand. After 25-30 minutes there Millions of copies of the desired DNA.16–20

Basic components of PCR
ℵ Complementary DNA (cDNA) or DNA template which contains the DNA fragment to be amplified.
ℵ Two primers that are used to amplify the DNA fragments.
ℵ Taq polymerase which copies the regions to be amplified.
ℵ Nucleotides are used for new DNA polymerization.
ℵ Buffer provides a suitable environment for DNA polymerase.
ℵ Thermal cycler (A machine that is used for maintaining the temperature during the reaction)
Primers
Primers are short, artificial DNA fragments not more than fifty usually(18-25bp) nucleotides that are corresponding to the start and end of the DNA fragment to be amplified. They anneal to the DNA fragment at the initial and endpoints where the DNA polymerase enzyme attaches and begins the synthesis of new DNA strands. DNA fragments that are to be amplified are determined by selecting the Primers. The PCR cycle consists of twenty to thirty-five cycles each consists of three main steps namely Denaturation, Annealing, and Extension. By using this technique, it is easy to find out the evolutionary relationship between the parents and off-springs. Agarose gel Electrophoresis is a procedure that consists of agarose gel in which DNA is injected into the gel and then apply the electric current to the gel. The result is that small DNA fragments move faster than the larger ones throughout the gel towards the positive current. The size of the PCR product can be measured by comparing it with DNA stepladder that contains DNA fragments with known size in the gel. By using the PCR technique hereditary diseases can easily check, handle, and cure significantly. Each gene can easily be amplified with known sequences to detect the mutations. Viral diseases can also be easily detectable by using this technique.21–24
Stages of PCR
There are mainly three main stages of PCR that are
ℵ Exponential amplification.
ℵ Leveling off stage.
ℵ Plateau
Exponential amplification
At this stage, only small quantities of DNA is required and assuming 100% reaction efficiency and at every cycle, the amount of DNA becomes doubled. This technique is very sensitive. Leveling off stage. At this stage DNA polymerase enzyme lose its activity and as consumption of reagent, such as primers or dNTPs are limiting them.
Plateau
At this last stage, no more component is produced due to disintegrating of regents and enzymes.
Steps of practical modifications of PCR. Main steps of practical modifications are given below:
ℵ Nested PCR
ℵ Inverse PCR
ℵ Reverse Transcription PCR
ℵ Asymmetric PCR
ℵ Quantitative PCR
ℵ Qualitative PCR
ℵ Touchdown PCR
ℵ Colony PCR
allele-specific PCR
ℵ Polymerase cycling PCR
ℵ Asymmetrical PC
A quantitative polymerase chain reaction is a PCR based laboratory technique which is also called real-time polymerase chain reaction which is used to amplify and quantify targeted DNA molecules. Usually, PCR takes place in a tube and when a reaction is complete the end product of the reaction can easily be visualized and analyzed by gel electrophoresis. However real-time PCR permits the analysis of products while the reaction is still in progress. This is achieved by different fluorescent dyes which react with enlarged product and it can be calculated by different instruments. This also facilitates the quantization of the DNA. As in real-time PCR procedure, exact quantity of PCR can easily be determined by this method or this technique is followed in Quantitative PCR (Q-PCR). In this method, quantitative amount of PCR, cDNA, and RNA can easily be measured. PCR is therefore used to measure whether a DNA sequence, several copies, or DNA is present in the sample. There is a key difference between the PCR and the traditional PCR is measuring the precise amount of Nucleic acid.
Application of PCR
The polymerase chain reaction has been a standard method that is used in all laboratories that carry out research on or with nucleic acid. It helps in exploring a number of growing diseases. The major advances in PCR applications are the use of reverse transcriptase to quantify the amplification of DNA in real-time and the extension of PCR technology for further scientific research. These techniques have provided greater information about diseases and scientific research by allowing the determination and quantificational changes in gene expression. PCR is used in research laborites in DNA cloning, southern blotting, recombinant DNA, and DNA sequencing technologies. In Microbiology, molecular biology labs PCR technique is used very frequently but in clinical microbiology PCR is very helpful because it is useful for analysis of microbial infections, and epidemiological studies. PCR is used in forensic laborites because a very small amount of original DNA is required that is even taken from single hair or droplet of blood. The PCR technique is very useful for the detection of a broad range of bacteria in CSF specimens have been reported. C. pneumonia is that organism who’s culturing and detection in clinical laborites is too much difficult but by using PCR techniques scientists can easily handle it this is the great achievement of PCR technology that it detected not only the genes of humans but also viruses, bacteria, and the detection of pathogens even it detects the number of pathogens in and outside the specimen, it also detects the amount of DNA of organisms in any foodstuff such as drinking water, foodstuff or the body fluids etc., it detects the exact quantity of microorganisms DNA. Many viruses contain RNA rather than the DNA so in such cases firstly viral genome converted to DNA for further processing by using RTPCR. PCR test is a very sensitive test about some viral infectious diseases such as human papilloma virus, herpes simplex virus, vericella-zoster virus infections. Other diagnostic uses of PCR including the genetic inherited and more infectious diseases such as AIDS, hepatitis, and cancer, these diseases are more dangerous and difficult to handle and to examine. ELISA test is useful for diagnosis of AIDS. So PCR can also be used to cure the diseases.
The advancement in science has changed our lives in positive ways that would have been unpredictable even half a century ago. Molecular methods have secured the phase in such a way by introducing the polymerase chain reaction technique that plays a positive role in human life as well as all living organisms. PCR is become very quickly an important tool in all scientific and medical laborites to improve human health and human life as well. PCR is a rapid technique with high sensitivity and specificity. It can also detect many rapid and mixed infections. It can also be used in many disease diagnosis and evaluation techniques that how much DNA of the organism is infected and how much quantity of harmful organism’s DNA in the specimen. So PCR has become very important and useful technique for medical, clinical, and research laborites.
None.
Author declares there is no conflict of interest.
None.

©2020 Nazir, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.