Journal of
eISSN: 2572-8466


Research Article Volume 2 Issue 4
Department of Plant Molecular Biology and Biotechnology, Navsari Agriculture University, India
Correspondence: Chaitanya Mogal, Department of Plant Molecular Biology and Biotechnology, Navsari Agriculture University, Navsari- 396450, Gujarat, India
Received: February 15, 2017 | Published: March 14, 2017
Citation: Mogal C, Singh D, Mehta A, et al. Isolation and biochemical characterization of phytase from different sources. J Appl Biotechnol Bioeng. 2017;2(4):152-155. DOI: 10.15406/jabb.2017.02.00039
Phytase (myo-inositolhexakisphosphate phosphohydrolase) catalyzes the hydrolysis of myo-inositol hexakisphosphate (phytic acid) to inorganic monophosphate and lower myo-inositol phosphates and in some cases to free myo-inositol. Two bacterial strains, one fungal strain and one plant source were screened for phytate degradation viz. Escherichia coli (ATCC 25922), Bacillus subtilis (ATCC 6633), Aspergillus niger (ATCC 16888) and Glycine max (JS-335). Among these, B. subtilis (ATCC 6633) strain was found to produce maximum clearance zone of 2.7cm on phytase screening medium containing sodium phytate as substrate, incubated at 37°C for 48hours. Phytase was isolated from B. subtilis (ATCC 6633), which was able to withstand temperature ranging from 40°C to 50°C, pH 4.0-6.0 and showed maximum activity at 50°C and 6.0 pH. However, phytase isolated from E.coli (ATCC 25922), A.niger (ATCC 16888) and Glycine max (JS-335) were able to withstand temperature ranging from 37.5°C to 45°C only and at pH 3.5-5.0.
Keywords: phytase, B. subtilis, pH; temperature, escherichia coli, inositol, a.niger, dephosphorylation
NA, nutrient agar; LB, luria-bertani; PDA, potato dextrose agar
Phytase, or myo-inositol hexakisphosphate phosphohydrolase (EC 3.1.3.8), was first discovered by.1,2 The principal end products of phytase action are phosphoric acid and myo-inositol, but the phosphatidylinositols representing various degrees of dephosphorylation from inositol hexakisphosphate to inositol are generated as intermediates. Phytate, an inhibitor of iron absorption, can be degraded by phytase. Phytase is an ester hydrolyzing enzyme with an estimated molecular mass of 35-700 kDa depending upon the sources of origin. Phytase genes have been isolated from plants,3 bacteria4 and fungi5,6 (Figure 1). Although phytase shows a potential to be utilized for phytate bioconversion, the enzyme activities and yields need to be increased to make them possible for industrial application. Therefore, it is important to isolate a variety of different microorganisms and their enzymes for phytate degradation.7 The pH optimum of phytases varies from 2.2 to 8. Most microbial phytases, especially those of fungal origin, have a pH optimum between 4.5 and 5.6. In contrast to most fungal phytases, A. fumigatusphytase has a broad pH optimum; at least 80% of the maximal activity is observed at pH values between 4.0 and 7.3. Some bacterial phytases, especially those from Bacillus, have a pH optimum at 6.5-7.5. The pH optima of plant seed phytases range from 4.0 to 7.5, most having an optimum between 4.0 and 5.6. The temperature optima of phytases vary from 45 to 77 °C. This paper describes a phytase isolated from different sources and their study of enzyme activity at different pH and temperatures.
The investigation was carried out using four organisms viz., Escherichia coli (ATCC 25922), Bacillus subtilis (ATCC 6633), Aspergillus niger (ATCC 16888) and Glycine max (JS-335). Escherichia coli (ATCC 25922), Bacillus subtilis (ATCC 6633) were grown at 37°C on Nutrient Agar (NA) and Luria-Bertani (LB) agar medium respectively. Aspergillus niger (ATCC 16888) was grown at 37°C on Potato Dextrose Agar (PDA) medium. The seeds of soybean variety (JS-335) were procured from Pulse Research Station, Navsari Agricultural University, Navsari and 9 days old seedlings were used for enzyme assay.
Biochemical characterization
Growth studies at different temperature and pH concentrations: To study the definite growth pattern of the isolates, first temperature and pH requirements were optimized for each strain.
Temperature optimization: Tubes of media were equally inoculated with fresh culture in duplicates and incubated in the range of 35-55°C (with an interval of 2.5°C). After 72hrs, growth on particular media was measured by spectrophotometer at 820 nm.
pH optimization: Tubes of media at different pH range 3.0 to7.0 (with an interval of 0.5 pH) were prepared in duplicates and after inoculation incubated at respective optimum temperature for each strain. After 72 h, growth on particular media was measured by spectrophotometer at 820 nm.
Screening for the efficient phytate degrading bacteria: Isolates that produced clearance zone were then plated on phytase screening medium containing calcium phytate as substrate. The clearance zone and colony diameter were measured after 2-5 days of incubation at 37°C.
Phytase enzyme assay: Bacterial and fungal strains were grown in 50 ml of liquid medium containing 0.1% sodium phytate, 1% peptone, 0.2% (NH4)2SO4, 0.05% KCl, 0.05% MgSO4.7H2O, 0.03% MnSO4, 0.03% FeSO4.7H2O, pH 7.5 in a 250ml flask and incubated at 50°C for three days on a rotary shaker at 200 rpm. Crude enzyme was harvested by centrifugation at 10,000g for 10 mins at 4°C and the clear supernatant was used as the source of extracellular phytases. For extraction of phytase from soybean, 1 g of seeds was ground to a fine powder in a prechilled mortar and pestle. To this powder, 100 ml of chilled extraction buffer [0.02 M Tris-HCl (pH 7.6) containing 0.1 % Triton X-100].The mixture was homogenized until a uniform slurry was obtained and then centrifuged at 12,000g for 30 min at 4°C. The supernatant was used for phytase assays.4
Sodium phytate was used as substrate for assaying the activity of phytase. Phytase activity was determined by measuring the amount of liberated inorganic phosphate. The reaction mixture consisted of sodium phytate (Hi media; 0.5% w/v) prepared in sodium acetate buffer (0.2 M, pH 5.5) and 0.2 ml of supernatant. After incubation at 50°C for 30mins, the reaction was stopped by adding an equal volume of 15% trichloroacetic acid. The liberated phosphate ions were quantified by mixing 100 μl of assay mixture with 900 μl of 1.0 M H2SO4- 10% ascorbic acid- 2.5% ammonium molybdate (3:1:0.1) (v/v). After 20 mins of incubation at 50°C, absorbance was measured at 820 nm.8
In the present study, two bacterial strains (B. subtilis and E. coli), one fungal strain (A. niger) and one plant source (G.max) were screened for phytate degradation. Among the isolates, B. subtilis (ATCC 6633) strain were found to possess maximum phytate degrading capacity which was identified by the presence of clearance zone in phytase screening medium containing sodium phytate as substrate. The result agreed with 1 who stated that the growth on plates was suggested to be easier than in a liquid medium for phytate degrading bacteria. The organisms obtained by spread plate method on phytase screening medium containing sodium phytate as substrate are shown in Figure 2. B. subtilis (ATCC 6633) strain was found to produce maximum clearance zone of 2.7 cm on phytase screening medium containing sodium phytate as substrate, incubated at 37°C for 48 hours. E. coli strain (ATCC 25922) and fungal strain i.e. Aspergillus niger (ATCC 16888) also produced clearance zone but the diameter of the clearance zone and growth of the organism was less compared to B. subtilis (Figure 3).
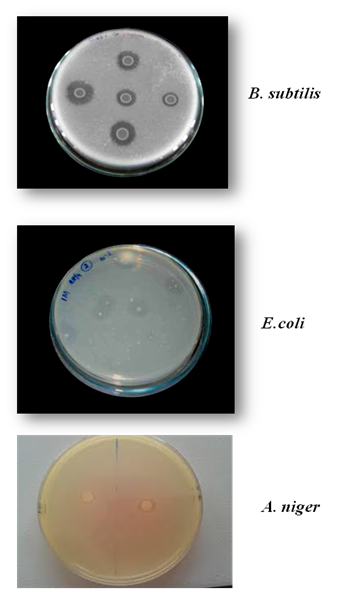
Figure 2 Clearance zone around the organisms when the sample was spread plated on phytase screening medium containing sodium phytate.
Phytase enzyme activity at different pH and temperature
After analysing the results on different pH it was observed that phytase enzyme isolated from B. subtilis posses maximum enzyme activity i.e. 4.20+0.02 U ml-1 at pH 6.0, where as E.coli, Aspergillus niger and Glycine max showed maximum enzyme activity i.e. 2.90±0.01, 2.90±0.01 and 2.75±0.01 respectively at pH 5.0 (Table 1 and Figure 3). After pH 6.0 there was reduction in enzyme activity of B. subtilis, where as in other organisms E.coli, Aspergillus niger and Glycine max after pH 5.0 the enzyme activity was reduced (Figure 3). Thus, based on maximum enzyme activity it may be said that phytase enzyme of B. subtilis may be more useful compared to other sources. Phytase enzyme of B. subtilis may perform better under acidic condition because at pH 5.0 also it showed higher activity i.e. 3.30±0.01 U ml-1 compared to other organisms. After analysing the results on different temperature it was observed that phytase enzyme isolated from B. subtilis posses maximum enzyme activity i.e. 4.20±0.02 U ml-1 at temperature 50°C, where as E.coli, Aspergillus niger and Glycine max showed maximum enzyme activity i.e. 2.67±0.01, 2.85±0.01 and 2.90±0.02 respectively at temperature 45°C (Table 2 & Figure 4). After 50°C there was reduction in enzyme activity of B. subtilis, where as in other organisms E.coli, Aspergillus niger and Glycine max after 45°C the enzyme activity was reduced (Figure 4). Thus, phytase enzyme of B. subtilis may be more thermostable compared to other sources.
pH |
B. subtilis |
E. coli |
A. niger |
G.max |
3 |
3.00±0.02* |
2.75±0.01* |
2.45±0.09* |
1.88±0.01* |
3.5 |
3.10±0.01* |
2.78±0.02* |
2.48±0.02* |
2.00±0.05* |
4 |
3.20±0.04* |
2.80±0.09* |
2.50±0.01* |
2.27±0.02* |
4.5 |
3.27±0.02* |
2.79±0.02* |
2.70±0.02* |
2.35±0.01* |
5 |
3.30±0.01* |
2.90±0.01* |
2.90±0.01* |
2.75±0.01* |
5.5 |
3.50±0.01* |
2.85±0.02* |
2.86±0.01* |
2.60±0.02* |
6 |
4.20±0.01* |
2.80±0.01* |
2.79±0.01* |
2.56±0.02* |
6.5 |
3.10±0.01* |
2.78±0.02* |
2.60±0.02* |
2.40±0.01* |
7 |
3.00±0.01* |
2.50±0.01* |
2.50±0.02* |
2.25±0.01* |
S.Em. |
0.059 |
0.062 |
0.059 |
0.058 |
C.D at 5% |
0.32 |
0.18 |
0.17 |
0.17 |
C.V.% |
3.27 |
3.43 |
3.31 |
3.25 |
Table 1 Phytase enzyme activity at different pH.
*The values are depicted as Mean ± S.D. of 5 independent observations.
Temp (°C) |
B. subtilis |
E. coli |
A. niger |
G.max |
35 |
2.00±0.01* |
1.99±0.01* |
1.85±0.02* |
1.95±0.01* |
37.5 |
2.25±0.02* |
2.10±0.02* |
2.22±0.01* |
2.00±0.01* |
±40.0 |
2.46±0.01* |
2.35±0.01* |
2.45±0.02* |
2.50±0.01* |
42.5 |
2.60±0.01* |
2.55±0.01* |
2.65±0.01* |
2.67±0.02* |
45 |
2.60±0.01* |
2.67±0.01* |
2.85±0.01* |
2.90±0.02* |
47.5 |
2.88±0.01* |
2.50±0.01* |
2.60±0.02* |
2.80±0.01* |
50 |
4.20±0.02* |
2.40±0.02* |
2.45±0.01* |
2.67±0.01* |
52.5 |
3.50±0.01* |
2.20±0.01* |
2.25±0.02* |
2.45±0.01* |
55 |
3.35±0.01* |
1.98±0.01* |
1.88±0.02* |
2.10±0.01* |
S.Em. |
0.085 |
0.064 |
0.079 |
0.091 |
C.D at 5% |
0.25 |
0.19 |
0.23 |
0.27 |
C.V.% |
4.91 |
3.68 |
4.64 |
5.24 |
Table 2 Phytase enzyme activity at different temperature (°C).
*The values are depicted as Mean ± S.D. of 5 independent observations.
Our finding suggests that phytase enzyme isolated from B. subtilis (ATCC 6633) has significant values which can be exploited for industrial production of phytase. Moreover, this enzyme can be used in the animal feed industry for improving the nutritional status of feed and in combating environmental pollution.
None.
The author declares no conflict of interest.

©2017 Mogal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.