Journal of
eISSN: 2572-8466


Research Article Volume 1 Issue 1
1Center for Bioengineering and School of Electrical and Computer Engineering, University of Oklahoma, USA
2Department of Pediatrics, University of Oklahoma Health Sciences Center, USA
Correspondence: Yuchen Qiu, Center for Bioengineering and School of Electrical and Computer Engineering, University of Oklahoma, USA, Tel 405-837-1998
Received: August 27, 2016 | Published: September 8, 2016
Citation: Qiu Y, Lu X, Tan M, et al. Increasing the efficacy of detecting residual cancer cells using a whole-slide image scanning method and a computer-aided detection scheme. J Appl Biotechnol Bioeng. 2016;1(1):1-6. DOI: 10.15406/jabb.2016.01.00001
Background and objective: The purpose of this investigation is to develop and test a new computer aided detection (CAD) scheme which is able to identify the residual cancer cells from the digitalized clinical specimens for the prognostic assessment of leukemia/lymphoma.
Methods: First, a whole slide image scan was performed by a commercialized fluorescent microscopic image scanner equipped with an objective 40× lens. Then, a computerized scheme was applied to detect and segment all clinically analyzable interphase cells depicted on the scanned images, as well as to recognize and count the independent FISH-probed signal dots within each interphase cell. Five pathological specimens were used to test the performance of this new scheme.
Results: The result shows that our scheme segmented and analyzed 4546, 3807, 2880, 2240, and 849 analyzable cells in five slides of different specimens including blood, bone marrow samples respectively, among which 334, 405, 178, 117, and 24 cells are detected by the scheme as suspiciously abnormal (or residual malignant) cells.
Conclusions: Comparing to the current visual detection method, the CAD scheme identified a much larger amount of FISH-probed cells. This investigation may help more sensitively detect residual cancer cells and improve the accuracy of prognostic assessment for leukemia/lymphoma patients in the future.
Keywords: interphase FISH, digital pathology, CAD, cancer prognostic evaluation, automated whole slide image scanning
Leukemia/lymphoma is a kind of malignant blood cancer with high heterogeneity.1,2 According to the WHO criteria, leukemia/lymphoma can be divided into the following categories:3
Although current average 5-year survival rate for all leukemia/lymphoma patients is ~50%, it varies significantly from one category to another4 as well as from the different individual cases in the same category, which can be attributed by the fact that the gene mutations are different in large number of subcategories.5,6 For example, in acute myeloid leukemia (AML) cases, the 5-year survival rates and tumor relapse rates vary from 15-70% and 33-78%, respectively.7 For the majority of leukemia/lymphoma patients, the “remission” is achieved by the initial therapeutic treatment in which no residual cancer cells are detected. However, in current clinical practice, only a small number of cells (e.g., ~200) are visually selected and analyzed by the technologists of cytogenetic laboratory under a microscope, which limits the sensitivity and statistical power of detecting the residual cancer cells. As a result, the residual cancer cells remain in many “remission” patients after the treatment but they may be missed in the cytogenetic lab tests. Accordingly, no further consolidation treatments will be applied to these "remission" patients, which may lead to cancer relapse. Therefore, identifying the residual cells is critically important for the post-remission evaluation of cancer prognosis and optimization of patient management.
Currently, in the cytogenetic laboratories, a number of technologies including chromosome banding analysis (CBA), interphase fluorescence in situ hybridization (FISH), and others, are applied for the leukemia/lymphoma diagnosis and treatment result assessment.8-10 Among these methods, the FISH technology utilizes specific fluorescent probes to detect and localize the interested deoxyribonucleic acid (DNA) segments on chromosomes.11 Comparing to the other two techniques, FISH technique demonstrates significant superiorities in discovering the cryptic aberrance or identifying structural or numerical abnormalities missed by CBA tests.10-13 As a result, current National Cancer Institute guideline recommends the incorporation of FISH test to the diagnostic work-up of leukemia patients.14
Despite of the advantages, the clinical potential of the FISH technology is limited due to the current visual FISH image examination method under a fluorescence microscope.15,16 In a cytogenetic laboratory, a technologist is required to visually select and analyze ~200 cells by switching between different fluorescent band-pass filters on the microscope.17,18 Previous studies have shown that selection and analysis of 200 cells could only achieve a very limited detection sensitivity with level p=0.1 (10% cells are malignant in the sample).19,20 In addition, visual selection and identification will inevitably generate bias and inter-observer variability,21,22 which may reduce diagnostic accuracy and consistency.23 For example, the technologists may not only miss the abnormal cells, but also sometimes purposely select more abnormal cells, eventually make normal vs. abnormal cells ratio completely differently from reality.
On the other hand, these studies19,20 also reported that in order to increase detection sensitivity to level p=0.01 (1% of cells are malignant), at least 4700 cells are required to be analyzed. Although one specimen slide may contain more than 4700 cell, it is impractical to detect and analyze such a large number of cells manually by the technologists in the cytogenetic laboratories. Hence, developing more sensitive and more efficient FISH cell screening and analyzing methods is critically meaningful to improve the accuracy for the diagnosis of the minimal residual diseases (MRD).20
Among many approaches to improve efficacy of detecting residual cancer cells, the development of the automated FISH image methods has been attracting extensive research interest since 1990s.24-26 Some of these studies utilized the commercialized image scanners to capture the images of the regions of interest (ROI) and then visually detect abnormal cells,27,28 while others developed and tested automated FISH signal dot detection and counting schemes.29,30 Due to the limited depth of field (DOF) of the microscope, these automated schemes require the fused FISH images captured at multiple focusing positions during the scanning,31,32 which is often inefficient and limits its potential in the busy clinical applications.
In this study, we developed and tested a new automated FISH image analysis method aiming to improve both the sensitivity and efficiency in the detection of residual cancer cells. For this purpose, the whole hybridization area of the samples was automatically digitized by performing an one-layer image scanning under a relatively low magnification objective lens (i.e. 40×, dry), which is able to yield a relatively large depth of field (DOF). A computerized FISH image processing and analysis scheme was then developed and optimized to detect the abnormal (cancer) cells. Five clinical pathological specimens were utilized in this preliminary study to test the feasibility of this new automated method. The details of this new automated method and experiments are presented as follows.
Clinical samples
In this study, we randomly selected five bone marrow (or blood) specimens from the patients who were undergoing the treatment of the diagnosed leukemia/lymphoma in the Health Sciences Center, University of Oklahoma (OUHSC). These patients are male, with an average age of 40.6 and they were diagnosed as CML, ALL, AML and NHL (Non-Hodgkin's lymphoma), respectively. In two of the five patients, translocations t (9;22)(q34;q11.2) and t(14;18)(q32;q21) were detected by the conventional chromosome karyotype examination. More translocations were identified on all the patients when using interphase FISH technique. The detailed diagnostic information of these specimens is illustrated in (Table 1).
Patient |
Gender |
Age |
Primary Disease |
Karyotype Results |
Interphase FISH Results |
1 |
M |
39 |
CML |
46,XY,t(9;22)(q34;q11.2)[11] |
(ABL1 con BCR)[122] |
2 |
M |
57 |
CLL |
46,XY[20] |
(CCND1 con IGHG1) [160] |
3 |
M |
25 |
AML |
46,XY[22] |
(PML con RARA)[71] |
4 |
M |
51 |
NHL |
46,XY,t(14;18)(q32;q21)[4] |
(IGHG1 con BCL2)[11] |
5 |
M |
31 |
AML |
46,XY[20] |
(PML con RARA)[7] |
Table 1 Patient information.
NHL: Non Hodgkin's Lymphoma, AML: acute Myeloid Leukemia, CML: Chronic myelogenous leukemia, CLL: Chronic Lymphoctic leukemia
Before conducting the interphase FISH investigation, these specimens were first preprocessed in our cytogenetic laboratory as routinely performed in current clinical practice. These samples were hybridized with BCR/ABL1, PML/RARA and IGHG1/BCL2 dual color (spectrum orange and spectrum green), dual fusion FISH probes (Vysis, Abbott Molecular Inc., Downers Grove, IL), to identify the translocations t(9;22)(q34;q11.2), t(15;17)(q24;q21.1) and t(14;18)(q32;q21), respectively.30,20 The preprocessing procedure is composed of three different steps. In order to detect the optical fluorescent property of leukemia/lymphoma cells, the cell nuclei were first extracted from the bone marrow (or blood) samples and spread on the surface of a slide in an area with a size of ~1.5cm × 1.5cm. The slide was then pretreated sequentially in the solutions of 2×SSC and 10% pepsin per 0.02 N HCI, at 37°C for 30 minutes. After that, the FISH probes were added and hybridization was performed overnight at 37°C. In the second day, post-hybridization procedure was conducted by treating the slides in 0.3% NP40 solution at 72°C for 1 minute and 0.1% NP40 solution at room temperature for 1 minute. The slide was finally stained with 0.5µg/ml DAPI (Vector Laboratories, Burlingame, CA).
Cytogenetic image acquisition
After sample preprocessing, each specimen was scanned by a commercialized digital fluorescent microscope (Eclipse Ni, Nikon Instruments, Tokyo, Japan), equipped with a digital scanning and camera control unit (Ds-Qi1, Nikon Instruments, Tokyo, Japan). The specimen slide was placed on a high precision x-y moving stage (MS-2000, Applied Scientific Instrumentation, Eugene, OR, U.S.A.) and an optical fluorescent filter (FITC-3540C, Nikon Instruments, Tokyo, Japan) was selected. In this proof-of-concept study, we only tested the concept of enabling to detect a much large number of cells by scanning the whole FISH slide using one color (Green) filter. During the scanning, we acquired the images containing both the interphase cell nuclei and the green color FISH-probed signals simultaneously. The auto-focusing operation was also applied to adjust the focal point with a distance of 1.5 mm. In order to improve image scanning efficiency and avoid repeated multi-layer scan, a 40× objective lens (dry, N.A. = 0.75) was used to replace the 100× oil emerged lens used in the manual examinations. The computed theoretical depth of field (DOF) of this 40× objective lens is 1.44 μm.33,20 According to our previous investigation, the captured cytogenetic images are visually analyzable if the cell is placed within three times of the theoretical DOF.34,20 Since the average thickness of bone marrow specimen is within the range of 4-5μm, a one layer scanning can cover most of the cell samples using this 40× objective lens.35,20
Cell segmentation
All obtained images were processed by a computer-aided image detection and analysis scheme. The scheme scanned the whole image to detect and segment the candidates for the potentially analyzable interphase chromosome cells. After the initial segmentation, all the separated regions were labeled and analyzed based on a set of decision rules to delete non-analyzable cells (i.e., clusters of cells with severe overlap, small and big debris regions), which have size outside the normal cell nucleus area range and/or irregular shapes. The remained candidate (cell) regions are considered as region of interest (ROI) and saved for conducting the following image processing.
Although the auto-focusing operation of the microscope was applied during the image scanning, some of the acquired cells may be off-focused or fuzzy. Therefore, our computerized scheme also automatically evaluates the image quality of each segmented cell candidate (ROI). In this study, two quality evaluation parameters, maximum intensity36,20 and Brenner gradient,37,20 were used in the scheme. The cell candidate is determined as analyzable (well focused state) if the maximum intensity is within the range between 80 and 200 and the Brenner gradient is between 20000 and 100000. These values are determined according to the clinical experience. Based on these two criteria, all the analyzable cells depicted on the scanned images are automatically stored in computer database and others are discarded.
Residual malignant cell identification
For each of these analyzable cell candidates, the scheme detected the FISH-probed fluorescent dots within the individual cell nucleus. Due to the small size of FISH signal dots and large intensity variation among different cells, our scheme sorts all pixels within the cell nucleus and separates the 50 brightest pixels from the background. A labeling algorithm was then applied to cluster the segmented pixels and create labeled dots. The algorithm applies two classification rules to finally detect the independent FISH signal dots. First, the isolated “dots” with the size smaller than one third of the maximal dots are considered as debris and deleted. Second, due to the operation control issues in FISH sample preprocessing step, one FISH signal dot may be smeared out and divided into two small dots with a close distance. The scheme used a distance based threshold to automatically determine whether two separated dots actually originate from the same targeted chromosome or not. According to the data reported in the literature, two dots are factually originated from one chromosome if their distance is smaller than approximately twice the average dot diameter.24
Finally, the scheme counted the detected FISH signal dots within each interphase cell nucleus. Although the detected and segmented cells may contain one, two, three, or more FISH-probed signal dots (in one color), the cells containing only one FISH-probed signal dot is not clinically useful. Missing the second FISH-probed dot can be typically attributed by the inevitable FISH processing error and/or limited depth of field of the objective lens.20,35 Therefore, the cells containing only one detectable FISH dot are discarded. The number of the cells depicting 2 (Normal) or 3 (Abnormal) visible (or detectable) FISH-probed dots was tabulated for comparison
Figure 1 shows an example image captured by the automated image scanner. It depicts a big cluster of many overlapped cell nuclei and several separated cells, some contain two FISH dots and some have three FISH dots. After the segmentation using our computerized scheme, the big cell cluster was deleted and two single separable cells were saved as analyzable ROI. Figure 2 demonstrates five examples of the segmented ROIs. Figure 2A & Figure 2B show one normal and one abnormal cell, containing 2 and 3 fluorescent dots, respectively. Both of these ROIs are captured at the in-focused state and the dots are very clear and bright. Figure 2C is an abnormal cell acquired at off-focused state but within the DOF range. The fluorescent dots are somewhat fuzzy, but can still be distinguished by our computerized scheme in detecting and counting the independent FISH signal dots. Figure 2D however, is captured out of the DOF, which is fuzzy and unrecognizable. Figure 2E shows a ROI containing debris. Both Figure 2D & Figure 2E were automatically discarded by our computerized scheme during ROI evaluation and FISH signal dot counting process.
Table 2 summarizes the experimental data generated from the five test specimens. For these specimens, our computerized scheme initially selected and segmented a number of 8093, 6675, 8354, 9570 and 4619 cell candidates (or ROIs) from the whole slide scanned images. After applying two cell selection criteria based on the maximum intensity and Brenner gradient, 4546, 3807, 2880, 2240 and 849 analyzable cells were automatically selected in these five specimens, respectively. Among these analyzable cells, our computerized scheme finally detected 334, 405, 178, 117 and 24 suspiciously abnormal cells with 3 FISH-probed signal dots in these five specimens, respectively. Comparing to the abnormal cells visually detected and reported by the cytogenetic laboratory technologists, the new method combines an automated whole slide image scanning microscopic device and a computerized FISH signal detection scheme, thus it enabled to detect substantially larger number of suspiciously abnormal (i.e. residual cancer) cells. For example, in specimen 4, the automated FISH analysis method detected over 10 times more abnormal cells than the subjective detection (117 versus 11).
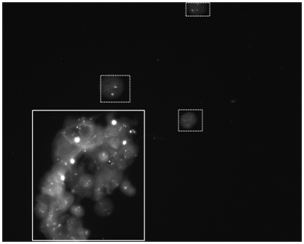
Figure 1 An example of FISH images acquired by the commercial automatic scanner. On this image, a typical cluster of cell nuclei and several typical isolated analyzable cell nuclei are marked in the real and dash boxes, respectively.
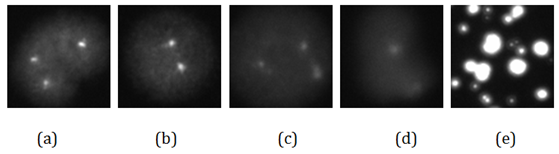
Figure 2: Five examples of segmented FISH ROIs.
(A) A normal cell, captured at in-focused state.
(B) An abnormal cell, captured at in-focused state.
(C) An abnormal cell, captured at off-focused state but within the DOF range.
(D) A cell captured outside the DOF range.
(E) An segmented false positive ROI containing meaningless information.
Specimen |
Detected ROI |
Analyzable ROI |
Normal ROI |
Abnormal ROI detected by the CAD scheme |
Visually Detected abnormal cells |
1 |
8093 |
4546 |
2081 |
334 |
122 |
2 |
6675 |
3807 |
1823 |
405 |
160 |
3 |
8354 |
2880 |
1711 |
178 |
71 |
4 |
9570 |
2240 |
1396 |
117 |
11 |
5 |
4619 |
849 |
521 |
24 |
7 |
Table 2 The number of the automatically detected normal and abnormal ROIs for the five tested specimens.
Unique characteristics
In this study, we demonstrated a fully-automated FISH image analysis method that combines a whole slide based fluorescent microscopic image scanning approach and a computer-assisted FISH signal detection scheme. The method has two unique characteristics. First, in image scanning, we tested a fully-automated, whole slide scanning method, which is implemented using a commercialized fluorescent microscope under a 40× objective lens. In the cytogenetic laboratories, the technologists usually use an oil-emerged objective lens with high magnification power (100×) to visually detect analyzable cells and count the FISH signal dots. Due to the small depth of the field (DOF) of the 100× lens, multiple layer scanning is necessary to cover the large sample thickness, which is inefficient for the busy clinical environment.34 Although one may use a low magnification lens (e.g., 10×) first to search for and detect the local regions of interest (ROIs) before the multiple layer image acquisition, this is a semi-automated approach and the entire procedure cannot be accomplished fully automatically. In order to address this technical challenge, we used a dry objective lens with 40× magnification power, which balances the scanning efficiency and spatial resolution. Our study demonstrated that this magnification power (40×) can significantly increases DOF of the system, and it is also sufficient for a computer-assisted scheme to detect FISH signal dots. As a result, a single layer whole slide image scan can be practically applied for the automated FISH image analysis purpose. When using a default auto-focusing function of the commercialized microscope, the scanning time of one slide takes from 1 to 1.5 hours in this study.
Second, we developed and tested a multi-stage computer-aided FISH image processing and signal detection scheme to detect and analyze all cell candidates depicted on a whole specimen slide. We applied two special cell candidate classification criteria (the maximum intensity and Brenner gradient) to distinguish between analyzable cells and other un-analyzable cells (i.e., debris and low quality or fuzzy cells). In addition, the FISH dot detection and segmentation algorithm was developed based on a unique adaptive threshold method. In order to identify and merge the separated dots originated from the same FISH-probed chromosome, the computerized scheme uses two simple classification thresholds based on the measured region size and distance to count the FISH signal dot numbers inside the nucleus. This study demonstrated that comparing to the conventional 200 cell visual analysis approach used in current clinical practice, our computerized scheme can detect and quantitatively analyze much larger number of cells. From the increased cell pool, the scheme also enables to detect much larger number of abnormal (cancer) cells.
Advantages and limitations
Comparing to the manual FISH image detection and analysis method used in the routine procedures of the cytogenetic laboratories to date, our computer-assisted automated FISH image analysis approach has several advantages. First, this new method can screen and process a much large amount (~3000) of interphase cells than the conventional visual screening, which may lead to detect the residual malignant cells with higher accuracy (p-level 0.01) than the manual screening.19,20 Second, it is a fully automated approach with higher efficiency than the existing automated scanning schemes, as the single position scanning method is applied during the specimen screening. Hence, it also has potential to help improve the efficiency of FISH image diagnosis. Currently, manual interphase FISH analysis is a time consuming process, which may take a technologist 30 to 60 minutes to visually investigate and analyze one FISH-probed slide in some samples.38 Although using our automated system may take approximately 1.5 hour to scan one specimen, this can be accomplished off-line and the clinicians may only need to spend several minutes to review the detected abnormal cells. In the residual malignant cancer cell detection, only a very small amount of cells will be finally determined as abnormal. For the conventional method, physicians have to spend most of the time on visually filtering out the normal cells under the microscope. Using this method, physicians can focus on analyzing the suspiciously abnormal cells selected by the CAD scheme, which may also potentially improve the diagnostic accuracy. In addition, the scanning time can also be significantly reduced by increasing the scanning speed or using the objective lens with lower magnifications (i.e. 20×). Third, this new technique may substantially reduce the cost of the cancer cell identification. The hardware of the automated system is commercially available with a price ranging from $50,000 to $150,000. Given that a typical life span of the automated scanning system is 10 years and the new method can significantly reduce the clinician's workload, the overall economic cost of using this new technique may be substantially cheaper than the conventional visual cell detection. Fourth, the automated scanning and computer-assisted quantitative image analysis may also help avoid the inter-observer variability and improve consistency of diagnosis.
Although the results of this preliminary study are encouraging, the study has a number of limitations. For example, the number of the test specimens is limited and only one channel of FISH probed signal was acquired for the verification of our new whole slide image scanning and computer-aided FISH-probed dot detection method. In addition, although our current computerized cell segmentation and FISH signal analysis scheme is able to detect and analyze much more FISH-probed cells than manual FISH image analysis method, a certain percentage of candidate cells are classified as un-analyzable. Hence, the computerized scheme has potential to be further improved to segment more analyzable cells (i.e., separating those slightly overlapped cells) in the FISH signal detection and analysis in the future.
This investigation demonstrated the feasibility of an efficient whole FISH-probed specimen scanning and signal identification method. Although there are several limitations, the automated FISH imaging method enables to detect and identify more FISH dot abnormal cells than current manual examination method. In summary, the new automated FISH image acquisition and analysis method may eventually help clinicians to increase the accuracy in detecting residual cancer cells for assisting the prognostic assessment of the leukemia/lymphoma patients.
This study is supported in part by the research fund from Peggy and Charles Stephenson Cancer Center, University of Oklahoma. The authors acknowledge the support of the Charles and Jean Smith Chair endowment fund as well. This paper has no conflicts of interest with other persons or organizations.
The author declares no conflict of interest.

©2016 Qiu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.