Journal of
eISSN: 2572-8466


Research Article Volume 7 Issue 6
1PhD Scholar, Department of Geography / Environmental Management, Faculty of Arts and Science, Nigerian Defence Academy, Nigeria
2Department of Biological Science, Faculty of Science, Nigerian Defence Academy, Nigeria
Correspondence: Ayodele A Otaiku, PhD Scholar, Department of Geography / Environmental Management, Faculty of Arts and Science, Nigerian Defence Academy, Kaduna, Nigeria
Received: June 08, 2020 | Published: December 28, 2020
Citation: Otaiku AA, Alhaji AI. Fungi consortia in situ biodegradation of xenobiotic, military shooting range, Kachia, Kaduna, Nigeria. J Appl Biotechnol Bioeng 2020;7(6):246-274. DOI: 10.15406/jabb.2020.07.00241
A major limitation of the white-rot fungus is its sensitivity during biodegradation of mixed matrix explosive pollutants and the scale of Kachia military shooting since 1967, Nigeria. The amplified 16S rRNA gene of each microbial isolate was processed for sequencing and characterization with Gene Bank database. Fungal species heavy metal reduction in increasing order of Aspergillus niger > Trametes versicolor > Rhizopus spp > Phanorochate chrysoporium > Penicillium spp were identified. The total explosive contents shows a significant difference for all locations in both dry and wet seasons (P<0.05) using Anova test. Microbial fungi consortium (MFC) bioremediate heavy metal significantly at 61.7% relative to isolated fungi species because of the lateral gene transfer/co-metabolism, where Trametes versicolor and Aspergillus niger act as gene mediators. MFC growth in 1% mineral salt medium munitions was significance than fungal species isolate. Deploying Myco Bio-augmentation / Phytoremediation/Biosimulation (Myco B-P-B) techniques to optimize the RDX and HMX characterized by a higher Nitrogen/Carbon ratio since fungi lack the beta-glucuronidase (GUS) gene to utilize carbon source directly. Pollutants bio-stimulation will enhances co-metabolism by MFC. Plant detoxification capabilities can be improved using fungi genes laccases and cytochrome P450 monooxygenase expressed effectively in plants using protoplast fusion.
Keywords: PC3R technology, explosives, enzymes, heavy metals, white rot fungi, peroxidase
co-metabolism, biofertilizer, bioremediation, entophytes, phytoremediation, tobacco, xenobiotic
myco B-P-B techniques and microbial fungi consortium
Xenobiotic waste streams constituents from explosives military shooting ranges operations globally results in the soil contamination are nitroaromatics and Nitra mines including TNT (2,4,6 trinitrotolueness), RDX (Hexahydro-1,3,5-trinitro-1,3,5-Triazine), HMX (Octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine), Picric Acid (2,4,6-trinitrophenol) Tetryl (Methyl-2,4,6 trinitro-phenylnitramine), PETN (Pentaerythritol tetranitrate), TATB (Triaminotrini-trobenzene). The impurities and degradation products from military shooting ranges includes: 2,4-DNT (2,4-dinitrotoluene), 2,6-DNT (2,6 dinitrotoluene), 4A-2,6-DNT (4-amino-2,6-dinitrotoluene), 2A-4, 6-DNT(2-amino-4,6-dinitrotoluene), TNB (1,3,5-trinitrobenzene), DNB (1,3 dinitrobenzene), NB (Nitrobenzene), Picramic Acid (2-amino-4,6-dinitrophenol) was reported.1–79 Microbial biodegradation using microbes is optimized when interact within their niche, under the most favourable conditions.80–279 Microbial bioremediation consortium techniques that occurs simultaneously during explosive biodegradation are: bio-stimulation, bio-augmentation, bio-accumulation was reported;356,280–372 bio-sorption136 the use of biofilms.318,320
Pollutants bioremediation involves a number of species of microbes having different mechanisms depending upon the environmental factors and the nature of chemicals 45 either to degrade or to remove the toxic contaminants from the environment. Elevated ambient atmospheric temperature (a climate change factor) may have impacts on the microbial activity,214 especially on the upper layers of soils. Explosives which are sufficiently sensitive to be detonated by a local hotspot or shock are called primary explosives (lead azide and lead styphnate) secondary (TNT, RDX) explosives less sensitive materials, which require an explosive shock wave for detonation can use primary explosives for detonation.117 Soil contamination by energetics at manufacturing sites, conflict areas, and military ranges is an international concern. Energetic compounds may enter the soil environment via numerous avenues31,24,333,215,216 including the following:
RDX is reportedly the most important military high explosive currently in use, being about 40% more powerful than TNT. Approximately 85% of all landmines contain TNT.166 At elevated CO2 microbe’s exhibit increased ability to decompose soil organic matter where soil microbial community plays an important role in cycling of carbon.246 The microbial biodegradation processes, the challenges are:
A fluo-spot approach,382 which can be employed for in situ detection of trace amount of explosives. The fluorescence molecule used is 4-(dicyanomethylene)-2-methyl-6-(4-dimethylaminostyryl)-4H-pyran) (DCM). DCM is featured by its high fluorescence quantum yield (15% in solid state, measured over the DCM deposited on a polytetra-fluoroethylene film) and unique electronic push-pull structure. The sensor reaction mechanism push-pull structure is a widely used design strategy for organic fluorescent molecules, as both the fluorescence efficiency and wavelength are very sensitive to the position and ability of the electron withdrawing or donating groups, which in turn can be changed by reacting with the analyses species (Figure 1).185, 399
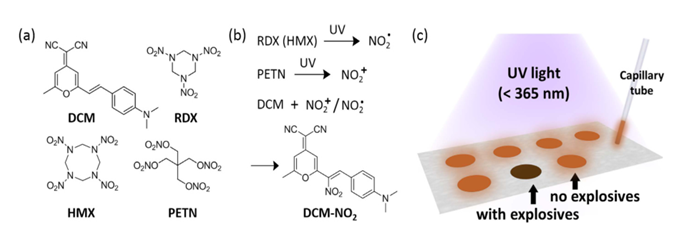
Figure 1 (a) Molecular structures of the fluorescence sensor, DCM, and the explosive compounds, RDX, HMX and PETN. (b) UV photolysis of RDX and PETN produces NO2· and NO2+ respectively; these reactive species can then be captured by DCM to produce the adduct DCM-NO2, leading to complete fluorescence quenching of DCM. (c) Scheme of the fluo-spot detection method adapted.382
Fungi biodegradation of TNT, RDX and HMX
Explosives environmental impacts on humans shows that RDX affects the central nervous system, and classified as class C carcinogens.289 HMX exists as α (orthorhombic), β (monoclinic), γ (monoclinic) and δ (hexagonal) forms, of which ‘β’ form is the least sensitive to impact and the most stable. HMX affects rats, aquatic organisms, bacteria, earthworm reproduction test and classified as Class D carcinogenic.289,348 White rot fungi are able to transform or mineralize TNT using nonspecific extracellular enzyme systems such as lignin peroxidase, manganese peroxidase, laccase. The findings asserted that micromycetous fungi like Alternaria spp., Aspergillus terreus, Fusarium spp., Mucor mucedo, Penicillum spp., Rhizoctoniai spp are able to transform TNT but do not mineralize it.409 Agaricus aestivalis, Agrocybe praecox and Clitocybeodora transform TNT, with degree of mineralization ranging from 5 to 15% reported.172,175 The white rot fungus P. chrysosporium degrade TNT with complete transformation 10 - 40% mineralization was reported.409 In a study reported313 that Phanerochaete Chrysosporium degraded RDX where it removed RDX (62 mg/L) from liquid medium containing glycerol as the main carbon source, Figures 2&3. Approximately 53% of the molecule was mineralized, and the major by product (62%) of the degradation was N2O. HMX was observed to be reduced from 61±20 mg/kg to 18±7 mg/kg during 62 days of incubation using P.chrysosporium. Under nitrogen-limiting conditions Phanerochaete chrysosporium mineralized HMX was reported113 After 25 days of incubation, 97% of HMX was removed via reduction with the accumulation of 4-nitro-2,4-diazabutanal was reported209 that the presence of glucose enhanced the degradation of HMX in marine sediment from a military UXO disposal and in which after 50 days, the HMX concentration in the aqueous phase (1.2 mg/L) was reduced by 50%. The disappearance of HMX was accompanied by the formation of a mononitroso derivative.
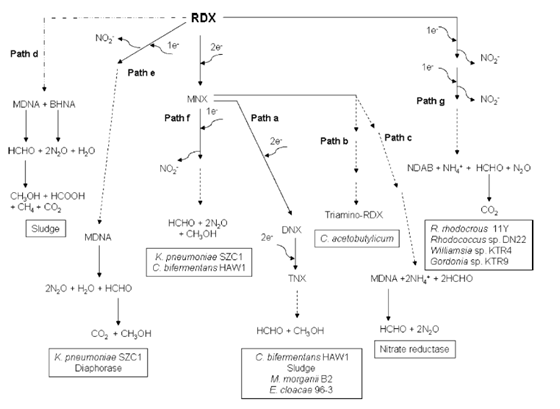
Figure 2 Proposed biodegradation pathways for hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX). Path a Reduction of RDX to nitres’ derivatives before ring cleavage.221 Path b Reduction of RDX to 1,3,5-triamino-1,3,5-triazine.408 Path c Reduction of RDX via Aspergillus niger nitrate oxidoreductase enzyme.34 Dashed arrows Multiple pathway steps based on hypothetical intermediates, which are not shown. Dashed plus dotted arrows hypothetical enzyme reactions.
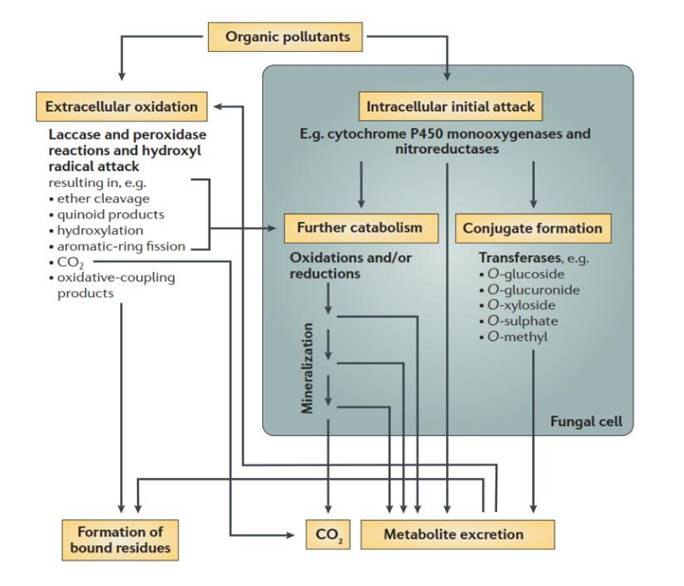
Figure 3 Principal methods used by fungi to degrade organic chemicals. Although fungi primarily
co-metabolize organic pollutants, they do grow volatile organics62, 271 ,169, 249,229 via pollutant attack may occur extracellularly or intracellularly adapted.147
Xenobiotic biodegradation treatments construct
Explosives biodegradation occurs in sequential incomplete process, potentially leading to the accumulation of toxic metabolites that can be further introduced into the food chain.366 This necessitate the convergence of bioremediation, phytoremediation and applied ecology xenobiotic degradation protocol called PC3R technology (Pollution Construct, Remediation, Restoration and Reuse) reported.256 Pollution construct (xenobiotic bioavailability in terrestrial environments for biodegradation) techniques (in situ or ex situ) Figure 4 soils/water contaminants for pesticides, herbicides, hydrocarbon, produced water, munitions, heavy metals and others for Remediation via microbial, phytoremediation and mineralization. Restoration ecosystems for zero pollutants footprints for sustainable re-generative Reuse of post treated soils site bio stimulated with biofertilizer for bioenergy crops (rice, soy bean or maize) and tobacco cultivation converted to biofuel production (ethanol). Rhizosphere remediation (plant-microbes interactions) by application for microbial biofertilizer (bacteria, fungi, etcs) to improves crop plant growth, soil ecosystem and biodiversity of the post-treated site.
Pollution construct
Laboratory studies reports that, most nitro-substituted explosives were found to be toxic for all classes of organisms, including bacteria, algae, plants, invertebrates, and mammals156,198,288,348 TNT and RDX are listed as ‘priority pollutants’ and ‘possible human carcinogens’ by the U.S. Environmental Protection Agency (EPA) reported.198,181 Phytoremediation, or the use of plants for cleaning up pollution304,308 metabolizing toxic pesticides.74,303 Environmental pollutants compounds may enter the environment during their production (wastewater lagoons), disposal (burn pits), storage, or usage (dispersed or unexploded ordnance), hydrocarbon and agriculture resulting in contamination of groundwater, surface water, marine, and terrestrial environments, and abiotic and biotic processes will influence the fate of explosive compounds.263,172 Physico-chemical properties of the compounds impacts on rate and extent of transport and transformation with environmental and biological factors including the presence and/or absence of explosives-degrading microorganisms Biodegradation pollutants construct solution become a requirement for the sustainable management of xenobiotic with converged technologies, Figure 4. Pollutants fate of compounds driven by: a). Processes that influence transport dissolution, volatilization adsorption b). Processes that influence transformation, photolysis, hydrolysis, reduction and biodegradation. Manufacture of explosives, testing and firing on military ranges, and decommission of ammunition stocks have generated toxic wastes, leading to large-scale contamination of soils and groundwater.146
Microbial remediation
Microbial bioremediation using microorganisms for the treatment of explosive-contaminated sites and range of contamination142 now the ‘top ten technologies for improving human health.83 Different classes of microorganisms, including bacteria and fungi, are capable of transforming energetic pollutants, which can be used as energy, carbon, and/or nitrogen sources reported by scholars.146,330,367,276 White-rot fungi that secrete powerful peroxidases that mineralization of TNT.367 P. chrysosporium is not a good candidate for bioremediation of TNT- contaminated sites containing a high concentration of explosives because of its high sensitivity to contaminants reported334 showed that the lignin peroxidase activity of P. chrysosporium is inhibited by the TNT intermediate hydroxylamino-dinitrotoluene. This is the prerequisite for the role of fungi consortium in xenobiotic bioremediation, Figure 3.
Phytoremediation
Transgenic plants305 exhibiting biodegradation capabilities of microorganisms bring the promise of an efficient and environmentally friendly technology for cleaning-up polluted soils.366 Phytoremediation of recalcitrant pollutants involve uptake, translocation, transformation and compartmentalization and sometimes mineralization.308 Plant roots secrete metabolites, which stimulate the growth of microorganisms in the rhizosphere, which in turn can degrade and mineralize the organic compounds. Plants lack xenobiotic degradative capabilities unlike bacteria of fungi. Therefore, introduction of genes for degradation of recalcitrant environmental pollutants from microorganisms or other eukaryotes will further enhance their ability to degrade/mineralize recalcitrant environmental pollutants. The MnP gene161 or the laccase gene from white-rot fungi has already been introduced into tobacco plants was reported (Figures 5–18).308 Hirai et al.,151 reported that laccase cDNA (scL) from white-rot fungus Schizophyllum was used as a model ligninolytic enzyme, efficient expression of scL and removal of a recalcitrant compound in transgenic tobacco plant by decreasing the CpGdinucleotide motif content in order to develop phytoremediation with a ligninolytic enzyme-producing transgenic plant. Phytoremediation merit over other remediation options includes: low installation and maintenance costs; and zero impact on the environment with carbon sequestration and biofuel production,93 Figure 16.
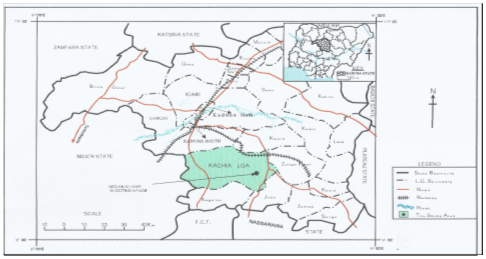
Figure 5 Map of Kachia within Kaduna state map, Nigeria showing study area (Nigerian Army Shooting Artillery, NASA Range).
Source: Geography Department NDA, Kaduna, Nigeria.
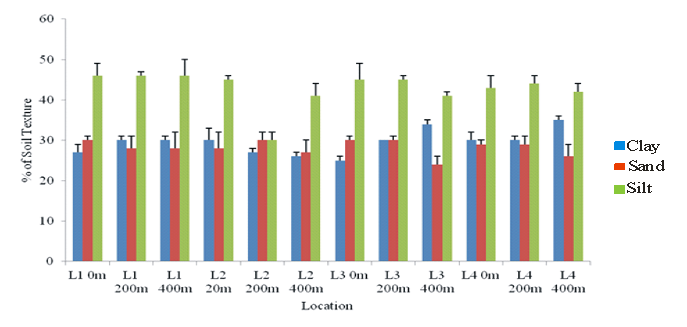
Figure 6 Percentage of Soil texture in various locations, Kachia, Nigeria. L10m Location (1) - zero metre, L1 200m Location 1 200 metre, Loc 1, 400 Location1 400 metre, Loc 20m - locations (2). 0 metre, Loc 2- Location (2).
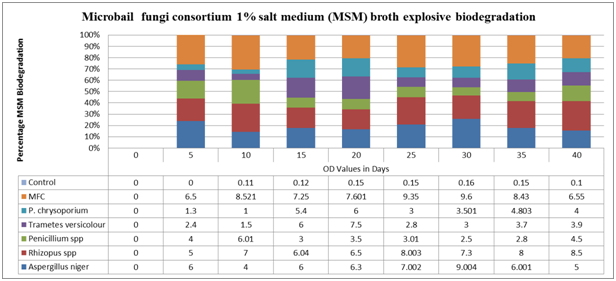
Figure 8 Growth Rate of (O.D Values) of Fungi in 1% Explosive Mineral Salt Medium broth.
*Means on the same column having different superscripts are significantly different (P<0.05) according to Duncan Multiple Range Test.
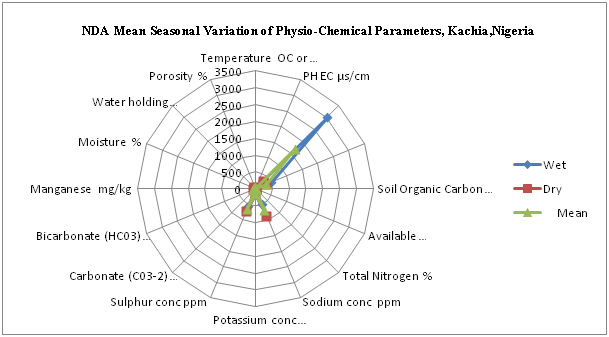
Figure 9 Mean seasonal variation of physcio-chemical parameters of composite samples of four shooting range locations.
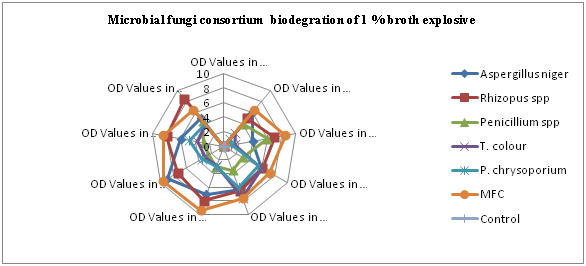
Figure 10 Shows the explosives fungal consortium isolates > Aspergillus niger >Trametes versicolor >Rhizopus spp > Phanorochate chrysoporium > Penicillium spp. identified. Similarly to the Gunasekaran et al.,135 reported that Rhizopus spp, Aspergillus niger, Trametes versicolor and Phanorochate chrysoporium as good mycoremediation potential for xenobiotic (Plate 1).
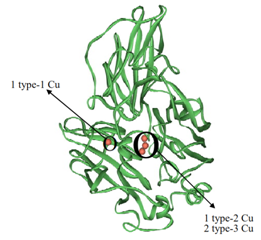
Figure 11 Ribbon representation of the X-Ray crystallographic structure of Trametes versicolor laccase adapted from Viswanath et al., 378 and www.chem.ox.ac.uk/icl/faagroup/fuelcell.html.
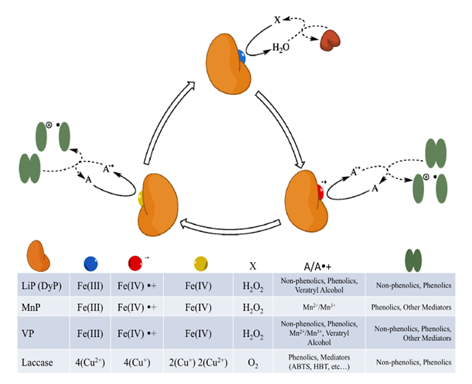
Figure 12 The catalytic cycle of ligninolytic peroxidases and laccases differ in their oxidizing substrate (X), their target reducing substrates/mediators (A) and their electron accepting metal co-factors (coloured circles). The peroxidases (LiP, DyP, MnP and VP) react with hydrogen peroxide to form oxo-ferryl intermediates (red and yellow circles), while laccases contain a four-copper active site that reduces oxygen to water to gain oxidative potential, adapted from Fisher et al.111
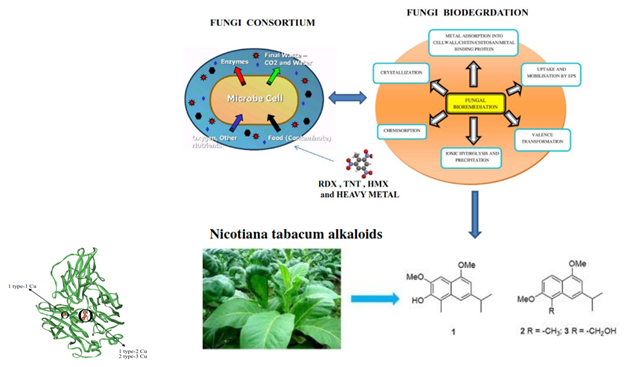
Figure 13 Illuminating the deconstruction of complex pollutants substrates requires many different types of enzymes and diverse chemical reactions within pollutants matrix simultaneously with resilience to environmental fluctuations by deploying microbial and phytoremediation combine techniques as affirmed by scholars.258,211
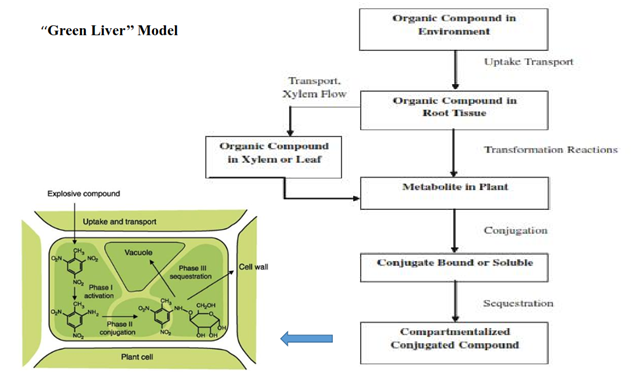
Figure 14 Green liver model for the metabolism of xenobiotic in plants and adapted from Burken et al.55
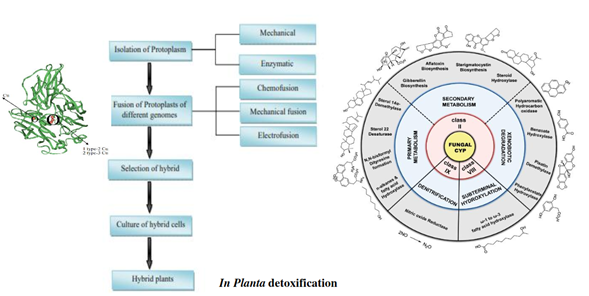
Figure 15 Schematic representation of production of hybrid plant via protoplast fusion.152 Representative scheme of functional diversification and classification of fungal CYP systems, adapted from Durairaj et al.,95,152 reports. Categorization of the functional properties of Class II CYP systems based on the primary metabolism, secondary metabolism and xenobiotic detoxification was adapted80 and ribbon representation of the X-Ray crystallographic structure of Trametes versicolor laccase modified.
www.chem.ox.ac.uk/icl/faagroup/fuelcell.html
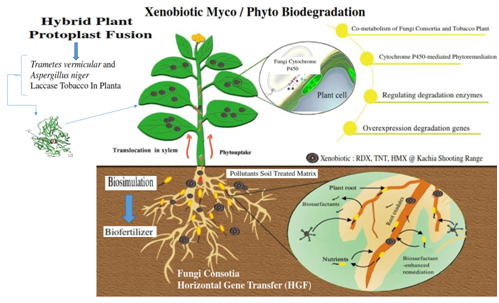
Figure 16 Potentially more cost-effective for in situ bioremediation, is the transfer of genes encoding microbial aerobic degradation pathways into transgenic plants tobacco (Nicotiana tabacum) will optimize the environmental risk to mineralization unidentified products during biodegradation, adapted Mo et al.238
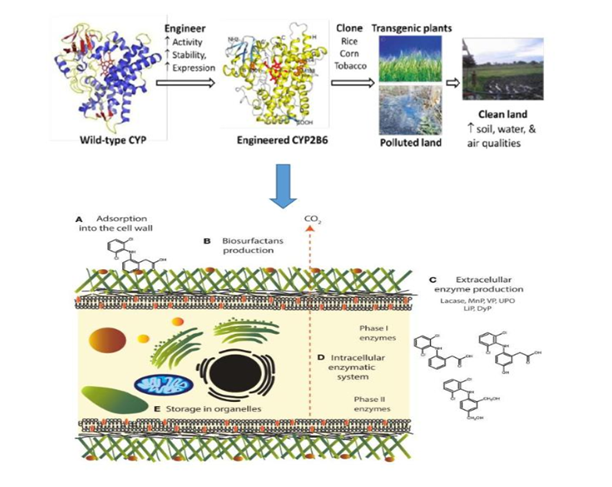
Figure 17 Using this genetic engineering, transgenic plants in which ligninolytic enzymes are expressed will be useful tool for phytoremediation of recalcitrant environmental pollutants adapted from Olicón-Hernández et al.249,305
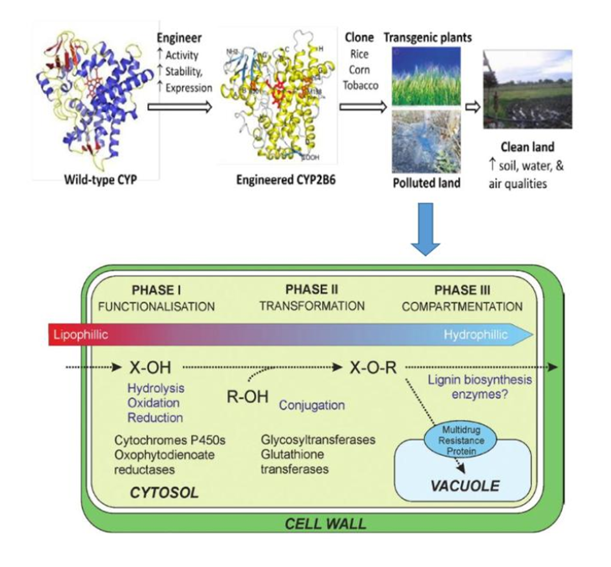
Figure 18 Classical pathway for xenobiotic detoxification in plants. In Phase I the foreign compound is functionalized, enabling Phase II, conjugation to sugars or glutathione, which facilitate Phase III, compartmentation to the vacuole or apoplast, adapted from Rylott et al.296
Plants lack the catabolic enzymes necessary to achieve full mineralization of organic molecules, potentially resulting in the accumulation of toxic metabolites.98 Microbes and mammals are heterotrophic organisms that possess the enzymatic machinery necessary to achieve a complete mineralization of organic molecules, complementing the metabolic capabilities of plants.98 The detection of oxidation and reduction metabolites, as well as large molecular weight products and non-extractable fractions51,120 suggests that plants metabolize explosives according to the ‘green liver’ model. 57,303 Like the liver of mammals, detoxification by plants typically involves three different phases: activation of the initial toxic compound (phase I), conjugation with a molecule of plant origin (phase II), and sequestration in plant organelles or structures (phase III). Plants via roots system have developed diverse detoxification mechanisms and have for long been recognized as capable of metabolizing organic compounds.74,303,320 Nitro-substituted explosives are efficiently taken up, Figure 13 and partially metabolized inside plant tissues. 55,93, 98
Restoration biofertilizer
Molasses is a very effective carbon source that enhances the TNT degradation rate significantly over other carbon sources46 and compost.396 Additionally, fungi can find application in phytoremediation if their potential genes of enzymes like peroxidases, laccases are expressed effectively in plants.328 Extensive reports on biofertilizer have revealed their capacity to provide nutrients to plants and consequently enhance crop yield.239,254,321 The goal of using such organic matter are twofold since it not only improves the quality and fertility of degraded soils but also simultaneously eliminates organic waste in a rational and environmentally friendly manner.148,390 Sustainable practices with the addition of biofertilizer255 (soil amendment) could be a useful tool for maintaining and even increasing the organic matter content for respiration in agricultural soils, thus preserving and improving soil fertility.310,311
Microbial communities contribute to fundamental processes that provide stability and productivity of agro-ecosystems.322 Soil biota affects ecosystem stability by regulating plant diversity, aboveground net primary production, and species asynchrony.401 The rhizosphere, which is the narrow zone of soil surrounding plant roots, can comprise up to 1011 microbial cells per gram of root100 and above 30,000 prokaryotic species that in general, improve plant productivity.227 The collective genome of rhizosphere microbial community enveloping plant roots is larger compared to that of plants and is referred as microbiome54 whose interactions determine crop health in natural agro-ecosystem by providing numerous services to crop plants viz., organic matter decomposition, nutrient acquisition, water absorption, nutrient recycling, weed control and bio-control.227,255
Reuse and mineralization
However, unlike bacteria and mammals, plants are autotrophic organisms that lack the enzymatic machinery necessary for efficiently metabolizing organic compounds, often resulting in slow and incomplete remediation performance.98 This led to the idea to modify plants genetically by the introduction of bacterial or fungi for complete xenobiotic biodegradations and mineralization.
The detection of TNT metabolites, such as aminodinitrotoluene and amino -dinitrobenzoate, suggests that plants can carry out both reductive and oxidative transformation of nitroaromatic compounds.276 Identified RDX transformation products include reduction derivatives, such as hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine,
and a range of breakdown products, such as 4-nitro-2,4-diaza-butanal, formaldehyde, and nitrous oxide.173,368 Another important barrier to field application of transgenic plants for bioremediation arises from the true or perceived risk of horizontal gene transfer to related wild or cultivated plants. Therefore, it is likely that the next generation of transgenic plants will involve systems preventing such a transfer, for instance by the introduction of transgenes into chloroplastic DNA or the use of conditional lethality genes.84
French et al.,116 designed the first transgenic plants specifically engineered for the phytoremediation of organic compounds by Doty94 tobacco plants (Nicotania tabacum) were modified by the introduction of a bacterial pentaerythritol tetranitrate (PETN) reductase, an enzyme involved in the degradation of nitrate esters and nitroaromatic explosives, and derived from an Enterobacter cloacae previously isolated from an explosive-contaminated soil.117 Meagher 226 report the rmineralisation of TNT by native plants is inefficient and generally incomplete.
To engineer plant tolerance to TNT, two bacterial enzymes (PETN reductase and nitroreductase), able to reduce TNT into less harmful compounds, were over expressed in N. tabaccum cv. ’Xanthi’. The two genes, onr (encoding PETN reductase) and nfsl (encoding nitroreductase), under the control of a constitutive promotor provided the transgenic plants with increased tolerance to TNT at concentrations that severely affected the development of wild type plants. Research schalars116,142,143,182 reported that, N.tabacum is able to extract metals only from the upper part of the contaminated layer (0.2 m) unlike Z. mays (maize) and S. viminalis which could extract metals from depths up to 0.75 m). Consequently, tobacco is suitable only for the phytoextraction of areas where the contaminated soil does not have a great depth.48
Rationale
Jimenez et al.,168 and Maruthamuthu et al.,216 applied selected microbial consortia, over single strains, for such biotechnological purposes presents important advantages. One of them is that microbial consortia are able to perform complex tasks181 that single strains. The reason is that the deconstruction of complex pollutants substrates requires many different types of enzymes and diverse chemical reactions, all at the same time or at short time intervals within pollutants matrix. Biodegradation is in phases, resilience to environmental fluctuations (resistance in variation substrate composition) and resistance to microbial invasion play major roles.211,258 White rot fungi can grow effectively under water stress conditions where no plant growth occurs.22 To optimize remediation approaches the development of genomes of specific white rot fungi like P. chrysosporium genome will result to the elucidation of specific cytochrome P450 monooxygenases, which may be differentially expressed in the presence of xenobiotic compounds.200 However, identification of specific interactions between microbes in a complex consortium is a difficult task, as it is highly likely that different interactions happen simultaneously.258 Application of synthetic microbial consortia could be a solution to this problem, as such consortia are usually composed of few well-identified organisms ex situ inoculant. At times, those species do not co-inhabit the same environment, but their metabolic activities may be combined.163 Determination of nutrient utilization, metabolite production, exchange of metabolites, production of signal molecules, spatial distribution, genome sequence and population dynamics are fundamental to enhance our understanding of the interactions, (Figures 4&16).
The knowledge could be applied to extend the stability of the microbial consortia and the process efficiency.126 The sheer scale of contamination of military shooting range in hectares,297means that while conventional remediation techniques may not be suitable, phytoremediation could be successfully applied to both remediate existing sites and to contain pollution from future use.297 RDX is readily taken up by plant roots and trans located to the above-ground tissues,206,376,377 but the in planta, Figure 14 (construct) degradation of RDX is low16,31,32,391 with much of the RDX likely to be released back into the ground following leaf abscission. The detoxification of RDX by endogenous plant enzymes involves the reduction to hexahydro-1-nitroso -3, 5-dinitro-1,3, 5-triazine and hexahydro-1,3-nitroso-5-nitro-1,3, 5-triazine,368 with nitrous oxide and 4-nitro-2,4-diazabutanal also detectable Photolytic degradation has also been reported.173
The logKow values are 0.90 for RDX235 and 1.86 for TNT and bioaccumulation has been reported for TNT transformation intermediates.23,179,202 By transferring microbial genes relaying xenobiotic-catabolism into selected plant species, detoxification, or, ideally, mineralization of organic pollutants can be achieved. Both RDX and TNT are toxic, organic pollutants which are recalcitrant to degradation in the environment.298 Additionally, fungi can find application in phytoremediation if their potential genes of enzymes like peroxidases, laccases are expressed effectively in plants.332 However, in toxic metal removal applications, it is important to ensure that the growing cells can maintain a constant removal capacity after multiple bioaccumulation–desorption cycles, and a suitable method is required to optimize the essential operating conditions. The situation demands a multi-prong approach including strain isolation, cell development and process development in order to make the ultimate process technically and economically viable.
A. niger could remove significant quantities of Cu and Pb from growth media but was less resistant against Cr.96 Multispecies consortia are considered more efficient over the monospecies culture due to greater resistance against environmental fluctuations and the metabolic relations among the member strains and the capabilities of such co-aggregates. Five different enzymes, lignin peroxidase, manganese peroxidase, laccase, cellulose and hemicellulase, were believed to be the most important catalysts in biodegrading process, and they always worked synergistically.66 Instead of depending upon single species, a better approach could be towards designing a consortium of strains having high metal biosorption, bioaccumulation and bioprecipitation capacities. The positive interaction between constituent species may also facilitate the survival of sensitive strains.47
Limitation of fungi biodegradation
Most of the studies dealing with microbial metal remediation via growing cells describe the biphasic uptake of metals, that is, initial rapid phase of biosorption followed by slower, metabolism-dependent active uptake of metals.88,123 However, in toxic metal removal applications, it is important to ensure that the growing cells can maintain a constant removal capacity after multiple bioaccumulation –desorption cycles, and a suitable method is required to optimize the essential operating conditions. The situation demands a multi-prong approach including strain isolation, cell development and process development in order to make the ultimate process technically and economically viable, (Figures 3&4). A. niger could remove significant quantities of Cu and Pb from growth media but was less resistant against Cr.96 However, until whole pollutant mineralisation, the use of different toxicity tests are needed to ensure the safety of the by-products formed171 and the rationale for PC3R technology. Whole genome sequence analysis can reveal the capability of fungi for multiple metabolic adaptations owing to diversified enzyme functions such as cytochrome P450 monooxygenase.159 Phytochelatin synthase (PCS) is an enzyme catalyzing the biosynthesis of phytochelatin from glutathione which protects cells against the toxic effects of non-essential heavy metals. The genome analysis of fungi is helpful in tracing such genes like pcs and studying their evolutionary aspects.316
A major limitation of the white-rot fungus is its sensitivity to biological process operations. The fungus does not grow well in a suspended cell system and enzyme induction is negatively affected by mixing actions and the ability of the fungus to effectively attach itself to a fixed medium is poor.77 The majority of the research on fungal performance has been conducted on autoclaved soil or on synthetic media. Although, the results show that the white rot fungus efficiently and successfully degrades highly toxic complex organic pollutants under these conditions, the results may not be as significant when they are grown under natural environmental conditions with variable native soil organisms, temperature, moisture and pH.149,277 One problem with field application deals with the strict growth conditions required for most white rot fungi. For example, P. chrysosporium has a high temperature requirement (30-37°C) for growth and ligninolytic enzyme production149 and like many white rot fungi, it has low competitive capabilities in the environment. Selection of fungal species with a competitive capability from nature and genetic engineering are needed for the future. The disadvantages of transforming filamentous fungi include changes in post-transcriptional treatment of recombinant proteins, resulting in low activity, defects in the morphology, low frequencies of transformation.381 Probably, the development of techniques of genome editing such as CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats) in fungi could solve these problems in a near future to optimize bioremediation process.
Study site
The study was conducted in the permanent military shooting/training range located at 5km east of Kachia town in Kaduna state, north central Nigeria. The range was established in 1965 and it covers an area of about 24.95 square kilometre that lies between longitudes 90 55’ N and 70 58’ E, with an elevation of 732 meter above sea level and the topography is undulating and the vegetation is Guinea Savannah, Nigeria (Figure 5, Table 1). The area where the munitions /explosive are fired (the impact areas) is a valley consisting of about four large rocks, where the fired munitions/explosives land and explode during military training. Five military exercises involving the deployment of explosives are carried out annually by the Nigerian Defence Academy (NDA) Kaduna, Nigerian Air force (NAF) Kaduna, Nigerian Army School of Infantry (NASI) Jaji, Armed Forces Command.
|
N/S |
Explosives |
Wet Season µg kg-1 |
Dry Season µg kg-1 |
|
1 |
TNT |
0.12–26.6 |
0.11–10.29 |
|
2 |
HMX |
1.43–68.19 |
0.39–35.67 |
|
3 |
RDX |
0.38–15.33 |
0.49–68.19 |
|
4 |
PETN |
1.17–7.12 |
0.39–4.38 |
Table 1 Explosives sites analyses of wet and dry seasons, Kachia, Nigeria
Sampling points
Four sampling points selected for the study are locations 1, 2, 3 and 4, Table 1, locations 1 and 2 are the twin smallest rocks closest to the road While locations 3 and 4 are much larger rocks heavily impacted by munitions/explosives between locations 1 and 2 is a flat ground where rain runoffs flow through to the stream in this site [map with global positioning system (GPS) coordinates, Figure 5. Locations 1 and 2 approximately 200 metres from the Plateau that is where the small arms are fired such as FN, Kalashinokov, Greenad, GPMG, SMG and Pistols. The soil in locations 1 and 2 is made of 50% silt and a flat ground with shrubs and drainage that flow through to the farm lands near the sites. Location 3 is approximately 9000metres away from the Plateau lies between 90 53’ and 44.71” N and 70 53’ 17.87” E. The impact area of locations 3 and 4 are mainly largely rocks containing high concentration of explosive due to the extensive use of bombardment by the artillery weapons, 155 mm nortwizer, and other heavy weapons while location 4 is ahead of location 3 is about 10,000 metres from the Plateau top where heavy weapons are fired too. The distance between locations 3 and 4 was covered with various shrubs and two major streams (Figure 5).
Sampling technique and soil treatment
Soil sampling: Sampling collected both dry and wet season. Four locations located within NASA (Table 1) shooting/training range Kachia were earmarked as sampling sites for this study using soil iron auger. Where 10 grams of soil sample (0-30cm in depth) with diameter of 9cm were collected from 3 different points within a location and harmonized to form a composite sample at various locations of the sites. All samples taken 2015 (June-August) and 2016 (February-March) were sieved using a 63 (106 m) mesh size laboratory sieve and then stored in black labelled polythene bags until for analyses. Samples for microbial analyses were kept in a cool box (site sampling) refrigerated with ice pack to retain the original microbial activities.
Soil sample pre-treatment
Sampling points were treated in the laboratory before digestion as executed. 10 grams of the soil sample was weighed into a clean dried beaker and put into an oven at about 100o C for one hour. The soil sample was then ground in a porcelain mortar with pestle and sieved through 250 µg mesh size to obtain a homogenous sample. The soil sample was stored in sterilized polythene bags, label and kept for next stage of pre-treatment. This procedure was repeated for all the collected soil samples. The ground soil samples were used for analysing heavy metal and explosives content for soil samples.165
Fungi species characterization
Preparation of media: Nutrient Agar - Nutrient agar (Antec /USA) by dissolving 28g of the Agar in 1 litre of distilled water in a conical flask. The conical flaks with the media were autoclave at 121°C at 15 pressure per square Index (PSI) for 15 minutes and cooled to 40°C before pouring or dispensing if a sterile petri dishes. MacConkey - MacConkey agar was prepared by dissolving 49.53g of dehydrated medium in 1000 ml distilled water in a conical flask. It was heated to dissolve the medium completely. The medium was sterilized by autoclaving at 15 1bs pressure (121°C) for 15 minutes and cooled to 45-50°C, mixed well before pouring into sterile petri plates.
Potato dextrote agar (PDA) Thirty nine (39) g of dehydrated medium suspended in 1000 ml distilled water was heated to dissolve the medium completely. It was then sterilized by autoclaving at 15 1bs pressure (121°C) for 16 minutes. Cooled ten percent (10%) of tartanic acid was added and mixed well to achieve pH 3.5 before dispensing into petri plates. The media were spread with the specimen soon after solidification of the media. Plates were incubated at 25-30°C in an inverted position (agar slide up) with increased humidity. Cultures were examined weekly for fungal growth and were held for 4-6 weeks.
Serial dilution
The sterile dilution blanks were marked in the following manner. 100 ml dilution blank was 102 ml and 9 ml tubes sequentially were 103, 104, 10-6 one gram of soil sample was weighed from each sample located and added to the 12-2 dilution blank and vigorously shaken for at least one minute with the cap securely tightened. All the 10-2 dilution was allowed to sit for a short period. The 1ml from this dilution was aseptically transferred to the 10-3 dilution was again transferred to the 10-4 dilution. The procedure was done to 10-5 and 10-6. A flask of nutrient agar from the 45°C water bath was especially pounded into each petri plates for that set. 15ml was poured enough to cover the bottom of the plate and mixed with the one ml inoculum in the plate. Each set was gently swirled on the bench so that the inoculum gets thoroughly mixed with the ager. All the plates were allowed to stand without moving so that the agar solidified and set completely. The plates were inverted and stacked into pipette carnisters and placed in the incubator or at room temperature until after 48 hours the same procedures were applied for MacConkey agar and Potato Dextrote Agar (PDA).16S ribosomal RNA and Polymerase Chain Reaction (PCR) for Amplification of Catabolic Genes Sequencing of the 16S ribosomal RNA (rRNA) gene and PCR based approaches. The concept of comparing gene sequences from microbial communities revolutionized microbial ecology.
Subsequently, a suite of molecular methods was developed that employ rRNA sequences10,128 referred to as rRNA. Medlin et al.,228 first described amplification of 16S-like rRNA from algae, fungi, and protozoa, and reports using 16S rRNA of bacteria and other eukaryotes soon followed.99,123,389,395 Bacterial diversity can be distinguished with 16S rRNA gene which is a universal marker for bacteria.352 Today mycological researches are also very keen for Deoxyribose nucleic acid (DNA) barcoding of fungal species. Examples of marker genes are present in multiple copies and contain conserved coding (small subunit, SSU and large subunit, LSU) as well as variable non-coding parts (internal transcribed spacers, ITS). Thus, they are useful to distinguish taxa at many different levels278 can be introduced for fungus taxonomical studies.67
Sequence-based classification and identification
Principle: Under the International Code of Nomenclature of Prokaryotes (ICNP)259 16S rRNA gene sequences are required for the description of new species, with a defined cut-off of 97% similarity between species.335,352,384 The connection of this DNA-based observation to its source organisms was made when researchers obtained rRNA gene sequences from previously cultivated fungi that matched the environmental DNA clade.292 One of the first such efforts was the Ribosomal Database Project (RDP)201,210 which initially consisted of nuclear small subunit (SSU) rRNA gene sequences from Archaea, Bacteria, and Eukarya. Recent additions to RDP include new databases for fungal ITS and large subunit (LSU) rRNA genes.72 The fungal LSU rRNA gene205 and ITS.270
The entirely on analyses of PCR-amplified nuclear rRNA genes, particularly the internal transcribed spacer (ITS) region, and they draw on incomplete taxonomic and functional databases. These are the so-called “dark taxa” 260 that reside in the International Nucleotide Sequence Database Collaboration (INSDC), with its three partners: Gene Bank at the National Centre for Biotechnology Information (NCBI). This is incorporated in a broader effort to add type material identifiers to the NCBI Taxonomy database, which allows use of BLAST and other tools to search multiple databases for reliable entries that are specifically tied to type material.107 PCR consist of an exponential amplification of DNA fragment and the principle is based on the mechanism of DNA replication in vivo, double stranded DNA is denatured to single stranded DNA, each single strand DNA is anneal by the forward and reverse primers of known sequence and elongated using tag DNA polymerase to produce copies of DNA template time for analysis as reported.190 The amplified 16S rRNA gene of each isolates were further characterized using gel electrophoresis. The amplified 16S rRNA gene of each microbial isolate was processed for sequencing and characterization.
The sequencing Kit (Applied Biosystems) with the Fungi Species Isolation of Genomic DNA. The genomic DNA of each fungus with observed remediation capabilities was isolated. 1ml of each fungal culture was pelleted by centrifuging at 12,000 rpm for 2 minutes; the pellet was treated with Iysis solution and proteinases K and incubated at 60°C for 30 minutes. DNA was extracted from each fungus precipitated with isopropanol by centrifuging at 10,000 rpm for 10 minutes, washed with 1 ml of a 70% (v/v) ethanol solution and dissolved in 0.1 ml of a T.E buffer. The purity and quantity of (DNA) of each sample was examined using UV absorption spectrum and agarose gel (1%) electrophoresis as described by scholars.174,347 Fungi species genes are present in multiple copies and contain conserved coding (small subunit, SSU and large subunit, LSU) as well as variable non-coding parts (internal transcribed spacers, ITS), Plate 1. Thus, they are product was analysed with ABI prism DNA sequence (ABI). The gene sequence of each isolates obtained in this study (Plate 1) were compared with known 16s rRNA gene sequences in the Gene Bank database as described by Jyothi et al.174
Procedure
Isolated explosive fungi DNA were amplified using catechol 2,3 dioxygenase gene primers. The genomic DNA, the primers and PCR master mix were added into a PCR tube. The tubes were spun to collect the droplets. The tubes were then inserted into the PCR machine while maintaining the regulation for initial denaturation (45 seconds at 94°C), annealing (1 minute at 55°C) extension (1 minute at 72°C) and final extension (10 minutes at 72°C). The amplified catabolic genes were resolved in 1.5% agarose gel electrophoresis stained with ethidium bromide and viewed under ultra-violet (UV light). The PCR reaction mixture containing 10 X PCR buffer, 25mm, magnesium chloride, 25mm dNTP’s, 10pm/uL primer concentrations and template DNA were used for the amplification of the 16S rRNA gene for each isolates. PCR conditions were optimized using lab net thermal cycler. The PCR program began with an initial 5-minuites denaturation step at 94°C and 35 cycles of 94°C for 45 seconds, annealing (1 minute at 55°C), 10 minutes extension step at 72°C.
Validated by comparison to ITS and LSU sequences within and among the same strains.349 As long-read sequencing technologies improve,191 it will become routine to obtain contiguous sequences containing ITS and its flanking LSU and SSU regions, which will facilitate integration of current datasets based on single markers. Fungal Genomes (1KFG) project,134 which is providing a platform for sharing protocols and for community networking and contributing to the transformation of fungal biology into a genomeenabled discipline.150 Genomic data from 1KFG and other projects may not have a major impact on species-level dentification, but they will be invaluable for predicting community function. Metagenomic binning makes it possible to assemble complete or near complete genomes from unculturable taxa while improved methods for isolating and growing previously uncultured microbes within their natural soil environments are opening possibilities to important new discoveries (Tables 2–4).203
|
Fungi isolates species |
Explosives |
Conditions |
References |
|
Rhizopus spp |
TNT |
TNT disappearance in malt extract broth |
Otaiku&Alhaji253 |
|
Electrophilic attack by microbial oxygenases |
Rylott&Bruce297 |
||
|
P. chysosporium spp |
TNT |
Mineralization to CO2 by mycelium and 90% removal of TNT |
Fernando et al.,1 19, Rh) et al.,280 Sublette et al.340 |
|
Mineralization to CO2 by spores |
Spiker et al.,331 Donnelly et al.89 |
||
|
Mineralization to CO2 by mycelium |
Michels and Gottschalk,230 Huang and Zhou155 |
||
|
Mineralization correlated with appearance of peroxidase activity |
Sublette et al.,340 Stahl&At336,337 |
||
|
Mineralization to CO2 in soil-corn cob cultures and stable |
Fernando and Aust,119 Yinon and Zitrin403 |
||
|
Compost microbes |
TNT |
Biotransformation by Thermophile microorganisms |
Kaplan and Kalpan,1 78 Isbister et al.160 |
|
Transformed ring-[ 14C]-labeled TNT,humification reactions |
Binks et al., 35 William et al.,396 |
||
|
P. chrysosporium spp |
RDX |
Biotransformation under nitrate-reducing |
Freedman&Sutherland114, Alc et al.8 |
|
Reduction by nitrogen-limiting conditions |
Fernando et al.,109 Fernando et al110 |
||
|
Reduction by sulfate-reducing condition |
Boopathyet al.,43 McCormick et al.221–223 |
||
|
Methylenedinitramine formation under nerobic condition |
Hawari et al.,114 Kitts et al.187 |
||
|
Aerobic denitration of RDX and highly mobile in soil |
Fournier et al.,113 ; Clausen et al.,70 Alaviet al.4 |
||
|
Reduction under methanogenic conditions |
Boopathyet al.,44 Kaplan177 |
||
|
Reductions lead to destabilization, ring cleavage, |
McCormick et al.,223 Clausen69 |
||
|
RDX |
Final products may include methanol and hydrazines |
Hawari et al.,146; McCornkk et al .,222 |
|
|
White rot fungus |
RDX |
Coagulation in contaminated water by nitrate reductase |
Sullivan et al ,343; Bhushan et al.,34 |
|
TNT |
Treated TNT in wastewater and achieved 99%. degradation |
Huang and Zhou155; Bennett25 |
|
|
TNT |
First step was degradation to OHADNT and ADNT, |
Aken et al.,3 Stahl&Aust337 |
|
|
Second step was to DANT including HMX and RDX. |
Axtel et al.,14 Stahl&Ast337 |
||
|
Aspergillus niger spp |
HMX |
Sequential denitration chemical substitution |
Urbanski,361 Kaplan177 |
|
Low water solubilitIity in anaerobic conditions |
Kaplan,176 McCormk et al.,223 |
||
|
PETN |
Mono-nitrated pentaerythritol to un-nitrated pentaerythrito products |
Williams and Bruce395 |
|
|
Mineralization |
PETN |
Biodegraded by sequential denitration to pentaerythritol |
Binks et al,35; Bait & Aust21; de Boer et al.,85 |
|
P. chrysosporium spp |
PETN |
Nitroglycerin, or glycerol trinitrate, degraded co-metabolism by sequential removal of nitro groups |
Wendt, et al.,386; Barr and Aust21 ;Stahl et al.,338 |
|
|
Angermaier et al.,12; Angermaier and Simon13 |
||
|
White rot fungus |
Explosives |
Dissolution rates: TNT > HMX > RDX>PETN |
Townsend and Myers,353 Brannon and Pennington49 |
|
|
|
Degraders glycerol trinitrate and isosorbide dinitrate : nitrate esters |
White & Snape392 |
Table 2 Fungi isolates implicated in biotransformation and mineralization. Kachia, Kaduna, Nigeria
Physio-chemical properties of the military firing ranges
The temperatures of 22-34°C during the dry season and 27.16°C during the wet season were recorded respectively, Table 4. The values obtained fell within the acceptable Federal Ministry of Environment (FMENV), Nigeria limit of <40. Similarly the pH of 6-8 has been suggested to be appropriate for microbial bioremediation.357 In the current investigation, the soil pH range for all the soil samples falls well within the range suggested by US-EPA357 and range of 27-35°C similar to the report of Jenkins et al.,166 on characterization of explosives contamination at military firing ranges with soils characterization of silt, clay and loamy, (Table 3&Figure 6).
The increased temperature obtained during the dry season can be attributed to the high concentration of explosives due to the frequent exercises by the personnel of Armed forces of Nigeria, Table 4 (Appendix 2). Compared with the wet season that the temperature is less as a result of rain washing away heavy metals and explosives drown the streams and soil porosity, Figure 6. The high value of electric conductivity (EC) recorded during the wet season is in agreement with the finding of Usman et al.,258 on their physico-chemical of the soils samples within the Federal ministry of environment, Nigeria ( FMENV) and World Health Organization (WHO) permitted limits of 500/µ5/cm and Penington et al.,263 within the value of 2000 mg/L which was contrary to the finding values were within 2400 -5900 mg/L and is in agreement with Usman et al.,258 the high values of Total Dissolve Solid (TDS) would be due to the deposit of organic and inorganic matters necessitated by the military activities.
It was revealed that RDX had the highest concentration of 68.19 µgkg-1 in dry season compared to wet season that was not very significant, Table 1 using the Anova test for total explosive contents shows a significant difference for all locations in both dry and wet seasons (P<0.05). In zero metre of the objective TNT, RDX and HMX were consistently found at the highest values of 49.39 mg/kg, 68.19 mg/kg and 35.67 mg/kg, respectively (location 4). An impact area and beyond within the hand grenade range, a 105 mm howitzer firing point and the heavy artillery and mortar range that had been analysed by GC-ECD. TNT concentrations ranged from 0.11 to 49.39 mg/kg in dry season and 0.12 to 26.6 mg/kg wet season. Among all the concentration of explosives both in dry and wet season RDX recorded the highest concentration of 68.19 mg/kg in Table 4 (Appendix 2) implied that Kachia shooting range consists of RDX and affirmed by Lachance et al.,198 with potential for carcinogen and the toxicity impacts on the ecosystem, (Table 2).
Myco-remediation of heavy metals: microbial fungi consortium (MFC)
Fungi role as primary and secondary decomposers in these classic "cycles" of nature.6,60,76,115 and spent fungal biomass from industrial fermentations is an available resource for the concentration of heavy metal contamination.121,122,293 Fungi myco-remediation mechanism are:
Although fungi participate in all three strategies, they are often more proficient at co-metabolism and bioaccumulation than at using xenobiotic as sole carbon sources. "Co-metabolism refers to any oxidation of substances without utilization of the energy derived from the oxidation to support microbial growth.157
The phenomenon has also been called “co-oxidation" and or "gratuitous" "fortuitous" metabolism. Many biochemists dislike the term.127,158 Nevertheless, it has become well entrenched in the literature. A second meaning of co-metabolism is to describe the degradation of a given compound by the combined efforts of several organisms pooling their biochemical resources for mutual efforts,82 Figures 7. Fungal isolates showed very good activity in the reduction of heavy metals when observed. Lead concentration was reduced by Aspergillus niger to 20%, Rhizopus to 31.5%, Penicillium spp to 25%, Trametes versicolor to 54.5%, P. chrysporium to 57% and Microbial fungi consortium (MFC) to 61.7%. Cadmium was reduced by A. niger to 16.5% Rhizopus spp to 17.5%, Penicillum spp to 15.3%, T.versicolor to 24.8%, P. chrysoporium to 20.5% and MFC to 100%. Zinc was reduced by A. niger to 32.49%, Rhizopus spp, to 38.8%, Penicillium spp to 34.2%, T. versicolor to 32.8%, P. chrysoporium to 37.85% and MFC to 39%.
Cobalt was reduced A. niger to 100%, Rhizopus spp to 19.7%, Penicillum spp 21.8%, T. versicolor to 100%, P. chrysoporium to 100%, and MFC to 100%. Copper was reduced by to 22.1%, Rhizopus spp to 6.5%, Penicillium spp 23.19%, T. versicolor to 34.5%, P. chrysoporium to 32.8% and MFC to 39%. Manganese was reduced by A. niger to 60.4%, Rhizopus spp to 7%, Penicillium spp to 16%, T. versicolor to 61.6%, P. chrysoporium to 63.3% and MFC to 68.2%. Nickel was reduced by A. niger to 17.1%, Rhizopus spp to 8%, Penicillium spp to 18.6%, T. versicolor to 26.8%, P. chrysoporium to 28.90% and MFC to 31.3%. Chromium was reduced by A. niger to 14.8%, Rhizopus spp to 5%, Penicillium spp to 10.9%, T. versicolor to 43.2%, P. chrysoporium to 41.7% and MFC to 45.5.2% while. Arsenic was reduced by A. niger to 13.3%, Rhizopus spp to 14%, Penicillium spp to 19.4%, T. versicolor to 19.8%, P. chrysoporium to 19.8% and MFC to 20.1%.
The best fungal isolates for heavy metal reduction in increasing order were Aspergillus niger > Phanorochate chrysoporium >Trametes versicolor > Rhizopus spp > Penicillium spp while, in the overall, the mixed culture consortium are the highest percentage reduction of each heavy metal analysed in this study as a result of co-metabolism and bioaccumulation as reported by Dagley.82 Anova test for total explosive contents shows a significant difference for all locations in both dry and wet seasons (P<0.05), Appendix 1. Fungi pattern of growth and development is less predictable with fixed into developmental pathways and affirmed by the report of Robson et al.,287 who reported that the life cycle of a fungus is unpredictable and flexible. The growth phases demonstrated by the fungi identified in this study show that the pattern of growth (Figure 8) displayed must have responded to the fixed nutrients of mineral salt medium supplemented with 1% munitions .The stages of growth are typically of any organism growing in a fixed quantity of nutrient. Every single isolate of fungus grew rapidly in the beginning that is, the exponential phase. This could be attributed to their previous association with munitions/explosives contaminated environment (Figure 10) and impacts of soil matrix on fungi physiology, Figure 11.
Appendix 1
|
Parameters |
Unit |
Wet |
Diy |
Mean |
|
Temperature |
OC or OF |
27.16 |
22.71 |
24.94 |
|
PH |
EC µs/cm |
6.07 |
2.31 |
4.19 |
|
Electric Conductivity (EC) |
EC µs/cm |
3011 |
312 |
1661.5 |
|
Total Dissolved solutes (TDS) |
(mg/L) |
513.1 |
347.7 |
347.7 |
|
Soil Organic Carbon (SOC) |
% |
5.11 |
5.76 |
5.44 |
|
Available phosphorus |
(mg/kg) |
4.7 |
4.9 |
4.8 |
|
Total Nitrogen |
% |
9.32 |
9.72 |
9.52 |
|
Sodium conc |
ppm |
498 |
863 |
680.5 |
|
Potassium conc (ppm) |
ppm |
169 |
162 |
165.5 |
|
Sulphur conc |
ppm |
560 |
724 |
642 |
|
Carbonate (CO3-2) |
mg/l00g |
0.22 |
0.35 |
0.285 |
|
Bicarbonate (ITCO3) |
mg/l00g |
1.11 |
1.68 |
1.4 |
|
Manganese |
mg/kg |
75.01 |
75.01 |
79.1 |
|
Moisture |
% |
59.56 |
59.56 |
35.87 |
|
Water holding capacity |
% |
59.56 |
51.12 |
55.34 |
|
Porosity |
% |
56027 |
69.39 |
62.83 |
Table 3 Mean seasonal variation of physcio-chemical parameters of composite samples of four firing rae locations (12 different metres)
Appendix 2
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX µg Kg-1 |
RDX µg Kg-1 |
PETN µg Kg-1 |
|
Wet |
Zero M |
L1 |
0-30 |
2.96 |
10.39 |
0.38 |
BDL |
|
Wet |
Zero M |
L2 |
0-30 |
2.1 |
7.12 |
BDL |
BDL |
|
Wet |
Zero M |
L3 |
0-30 |
26.6 |
29.6 |
15.33 |
BDL |
|
Wet |
Zero M |
IA |
0-30 |
1.34 |
2.21 |
7.42 |
2.07 |
|
Explosives µg Kg-1 |
|||||||
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX µg Kg-1 |
RDX µg Kg-1 |
PETN µg Kg-1 |
|
Wet |
200M |
L1 |
0-30 |
0.12 |
1.43 |
0.449 |
0.17 |
|
Wet |
200M |
L2 |
0-30 |
BDL |
6.31 |
BDL |
BDL |
|
Wet |
200 M |
L3 |
0-30 |
BDL |
BDL |
19.9 |
BDL |
|
Wet |
200 M |
IA |
0-30 |
0.19 |
BDL |
BDL |
1.39 |
|
Explosives µg Kg-1 |
|||||||
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX µg Kg-1 |
RDX µg Kg-1 |
PETN µg Kg-1 |
|
Wet |
400M |
L1 |
0-30 |
BDL |
12.4 |
0.67 |
1.15 |
|
Wet |
400M |
L2 |
0-30 |
BDL |
6.11 |
BDL |
7.12 |
|
Wet |
400M |
L3 |
0-30 |
0.39 |
BDL |
11.8 |
BDL |
|
Wet |
400 M |
IA |
0-30 |
BDL |
BDL |
12.73 |
0.71 |
|
Explosives µg Kg-1 |
|||||||
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX µg Kg-1 |
RDX µg Kg-1 |
PETN µg Kg-1 |
|
Dry |
Zero M |
L1 |
0-30 |
10.29 |
17.6 |
16.13 |
3 |
|
Dry |
Zero M |
L2 |
0-30 |
1.14 |
19.88 |
22.61 |
0.39 |
|
Dry |
ZeroM |
U |
0-30 |
16.31 |
33.07 |
23.14 |
BDL |
|
Dry |
Zero M |
IA |
0-30 |
49.39 |
35.67 |
68.19 |
1.6 |
|
Explosives µg Kg-1 |
|||||||
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX µg Kg-1 |
RDX µg Kg-1 |
PETN µg Kg-1 |
|
Dry |
200 M |
L1 |
0-30 |
0.11 |
1.6 |
15.11 |
BDL |
|
Dry |
200 M |
L2 |
0-30 |
0.13 |
13.31 |
2.69 |
BDL |
|
Dry |
200 M |
U |
0-30 |
2.05 |
BDL |
BDL |
BDL |
|
Dry |
200 M |
IA |
0-30 |
1.79 |
2.97 |
1.08 |
BDL |
|
Explosives µg Kg-1 |
|||||||
|
Season |
Distance Meter (M) |
Sites |
Depth (Cm) |
TNT µg Kg-1 |
HMX ig Kg-1 |
RDX ig Kg-1 |
PETN ig Kg-1 |
|
Dry |
400 M |
L1 |
0-30 |
BDL |
0.39 |
BDL |
BDL |
|
Dry |
400M |
L2 |
0-30 |
2.16 |
BDL |
BDL |
BDL |
|
Dry |
400 M |
U |
0-30 |
0.15 |
BDL |
2.38 |
BDL |
|
Dry |
400 M |
IA |
0-30 |
BDL |
0.69 |
0.49 |
4.38 |
Table 4 Mean seasonal variation of total explosives concentration in various locations of NDA shooting range Kachia, Kaduna state µg/ kg, Nigeria
N.B: TNT, 2, 4, 6 – trinitrotohience; RDX, royal demolition explosive; – 1,3,5 frinitro - 1,3,5 – trazinanze; HMX, 1, 3,5 tetranitor 1,3,5 – tetrazocalne high melting explosion; BDL; below detectable level
This is in conformity with Diaz & Massol-Deya,92 discovered that most of the fungal species were able to grow efficiently and appear concurrently in the environment as novel growth and energy substrate. Therefore, these fungi have a potential to degrade xenobiotic compounds (Figure 9). The growth pattern of fungus had different maximum growth peaks. This could be as a result of exhaustion of nutrients and release of toxic materials into the medium (transformation of waters explosives) and there is a need for improved biodegradation techniques with zero pollutants footprint (Figure 4). The growth pattern of fungi in 1% munitions/explosives and minimal salt broth is represented Figure 8.
Similarly, Gunasekaran et al.,135 identified Rhizopus spp, Aspergillus niger, T. versicolor and P. chrysoporium as good mycoremediation potential for xenobiotic pollutants (Plate1). The fungal consortium performs better than the single culture isolates of the fungi species which may be as result of a synergistic interaction of the fungal isolates. Similarly, Okerentugba & Ezeronye252 reported that a single microbe does not possess the enzymatic capability to degrade all or even most of the organic compounds in a polluted soil but mixed microbial communities possess powerful biodegradable potential because the genetic information of more than one organism is necessary to degrade the complex mixtures of organic compounds present in contaminated areas.
Fungi species (explosives by turbidometry) - the growth potential
Fungal celluloses provide a good example of the contrasting faces of a single enzymatic capability, example in the non-specificity of the white rot fungi is ideally suited to treating low concentrations of mixed wastes in a nutrient deficient habitat, complete pathways of degradation are more likely to occur through the combined effects of many organisms biochemistry. These include fungal amylases, glucoamyases, lipases, pectinases, and proteases27,132 and secondary products like LiP and MnP.145 Penicillium janthinellum degrade high molecular weight polycyclic aromatic hydrocarbons more efficiently than does either microorganism alone.40 The Figures 8 and 10 shows the optical density reading of biodegrading activity of each fungal isolate on 1% explosive mineral salt medium (MSM) broth (Basal salt medium). It was revealed that Aspergillus niger had a maximum growth peak at 9.004 on the 30th day as affirmed by similar work of Svecova et al.,344 Rhizopus spp had maximum growth peak at optical density 8.000 - 8.500 on the 25th, 35th and 40th respectively, Penicillium spp had the lowest growth peak at optical density 2.500 on the 30th day while the maximum was 6.010 on the 10th day. The maximum growth peak of Trametes versicolor was at 7.800 on the 20th day and the lowest growth peak was at 1.500 on the 10th day (Plate 1).
Phanorochate chrysoporium had the maximum growth peak at 6.000 on the 20th day while the lowest for P. chrysoponium was at 1.000 on the 10th day as a results of the impacts of lignin peroxidases genes [lipA, lipB, lipC, lipD, lipE, lipF, lipG, lipH, lipI and lip J ] as reported affirmed by scholar’s reports.214,409 Aspergillus niger had the best ability to degrade explosive while Penicillium spp had the least ability. The analysis of variance result shows that there is a significant difference in the overall growth pattern of fungi in MSM broth supplemented with 1% v/v explosive (P>0.05). The overall result also indicates that the growth rate increased significantly from the zero hour to the 5th day and beyond. Means on the same column having different superscripts are significantly different (P<0.05) according to Duncan multiple range test.
Furthermore, Fritsche et al.,118 demonstrated that manganese peroxidase and lignin peroxidase from P. chrysoporium and T. versicolor have the ability to oxidize or reduce cyclic nitromines. This result is in agreement with Sheremata & Hawari214 similar work revealed that P. chrysoporium, T. versicolor, or Aspergillus niger and Rhizopus spp, are capable of utilizing heavy metals (Figure 7) and TNT, RDX and HMX. Also, in this study, fungi pattern of growth and development is less predictable. Fungi were not fixed into developmental pathways. The life cycle of a fungus is unpredictable and flexible because the mechanism of action is co-metabolism and bioaccumulation as confirmed by Dagley.82 Complete transformation of TNT and 10 - 40% mineralization were discovered in Phanerochaete chrysosporium, a white rot fungi was reported.214,409 P. chrysosporium, a white rot fungus, was used in the degradation of RDX. It removed RDX (62 mg/L) from liquid medium containing glycerol as the main carbon source. Approximately, 53% of the molecule was mineralized, and the major by product (62%) of the degradation was N2O. P. chrysosporium mineralized HMX under nitrogen-limiting conditions. After 25 days of incubation, 97% of HMX was removed via reduction with the accumulation of 4-nitro-2,4-diazabutanal. Zhao et al.,409 reported that, the presence of glucose enhanced the degradation of HMX in marine sediment from a military UXO disposal and in which after 50 days, the HMX concentration in the aqueous phase (1.2mg/L) was reduced by 50%. The disappearance of HMX was accompanied by the formation of a mononitroso derivative.
MFC biodegradation significance
Fungal biodegradation mechanisms life cycle is unpredictable and flexible with intermediate bioremediation products which showed formation of MNX and ring cleavage and the subsequent formation of methylene dinitramine (MEDINA) by the white-rot fungus P. chrysosporium with RDX214 because the mechanism of action is co-metabolism and bioaccumulation as confirmed by Dagley.82 Nutrient availability is one important factor that plays a significant role in the rate of explosives biodegradation. Compared to other common pollutants, explosives, particularly RDX and HMX, are characterized by a higher N/C ratio. Although they may theoretically serve as sources of both carbon and nitrogen, only nitrogen is used by some organisms, and hence, another carbon source must be added9 and see Figure 4. Since denitration is frequently carried out by strains that are capable of utilizing the explosive compound as a nitrogen source, a negative response is often observed in microcosm experiments when external nitrogen sources (either ammonium or nitrate) are added, as these nitrogen-containing compounds may compete with degradation of the explosives for nitrogen.30,35,73,291, 243
There is a need for the development of ex-situ bio-augmentation products for the biodegradation of xenobiotic to assist mineralization because the additional nitrogen supports microbial growth without competing with the explosive compounds for metabolism of RDX as reported by scholars291,329,379 and TNT 41 nitrogen-bearing compounds are also potential inhibitors of enzyme expression, as presented by Nejidat et al.,243 who showed that ammonium and nitrite repress fungi lignin peroxidase. The mechanism by which the fungi mineralize the explosive is not known, but preliminary information about the process has been reported230,231 and mineralization by microbial consortium affirmed in similar work by Young et al. 404,405 employed a consortium from horse manure to successfully biodegrade RDX. Biodegradation rates are also influenced by the type of electron acceptors and electron donors. For example, the degradation of RDX, HMX and TNT was shown to be enhanced by additional hydrogen or hydrogen-producing electron donors under anaerobic conditions 1,2 and the degradation of HMX was enhanced by adding mixed electron acceptors to anaerobic microbial consortia.45 Redox potential is another important factor that not only plays an important role in dictating the mechanism of degradation, but also influences the actual biotic degradation rate. Biodegradation is generally enhanced under reduced conditions, and in saturated soils.272, 283,329
Laccase reaction mechanism
Laccases are of particular interest with regard to potential applications, because of their capabilities to oxidize a wide range of environmentally dangerous substrates. Greater attention on laccase, an eco-friendly enzyme and a green catalyst in recent past is generating information that appeared in a number of reviews.17,321,388 Some soil ascomycete species from the genera Aspergillus, Curvularia and Penicillium,19 Trametes versicolor,290,232 are some examples of basidiomycetes that produce laccases.378 Laccases from different sources displays a wide range of redox potentials. The T1 site of laccase of T. versicolor shows a high redox potential of 780-800mV,267 Figure 10 Expression of laccase in some fungi is regulated by nitrogen-limiting conditions102,186,262,268 while in others nitrogen sufficiency results in enhanced enzyme production. The effect of various nutrient nitrogen concentrations on expression of lcc genes at molecular level in T. Versicolor was examined.75 There was a direct correlation between concentration of nitrogen nutrient in growth medium and the level of lcc expression by T. versicolor. Copper is also often a strong inducer of laccase gene transcription, and this has been suggested to be related to a defense mechanism against oxidative stress caused by free copper ions. 75,86,108,119,204, 257,327 In addition to copper, other metal ions such as Mg2+, Cd2+ or Hg2+ can stimulate laccase expression.119,306,327 Figure 9 shows the explosives fungal consortium isolates impacts on biodegradation of munitions explosive because of laccase. The importance of adequate copper concentration for proper laccase folding was further corroborated by studies in which two genes related to copper-trafficking in T. versicolor were over-expressed in S. cerevisiae expressing T. versicolor lcc3 gene; the heterologous laccase production by S. cerevisiae was improved up to 20-fold,361 Figure 11. The effect was suggested to result from more efficient transport of copper to the Golgi compartment.361 Laccase biocatalyst protein engineering potentials in the development of robust, active and tailor made enzymes. In addition to substrate oxidation, laccase can also immobilize soil pollutants by oxidation, coupling to soil humic substances - a process analogous to humic acid synthesis in soils39 and an active biocatalyst for biofertilizer.255
Also, engineered to improve the efficiency of particular bioremediation processes like the T. versicolor in Aspergillus niger 38 and mediators to attack non-phenolics. Laccases are multi-copper oxidases produced by fungi, bacteria, plants and even insects that catalyse the one-electron oxidation of four equivalents of reducing substrate, with the corresponding four-electron reduction of atmospheric oxygen to water,97,68, 317 Figure 12. Fungal laccases are known to have higher redox potentials than bacterial laccases and are produced by a wide range of fungi. Biological role and significance of laccases in lignin degradation remains unclear like P. chrysosporium contains no laccase homologs in its genome, yet depolymerize lignin101,168,218 and see Figure 8. The application of selected microbial consortia, over single strains, for such biotechnological purposes presents advantages to perform complex tasks that single strains .The reason is that the deconstruction of complex substrates requires many different types of enzymes and diverse chemical reactions within pollutants matrix simultaneously with resilience to environmental fluctuations was reported258,211 and illustrated by Figure 12 (microbial and phytoremediation construct).
Application of synthetic microbial consortia could be a solution to this problem, as such consortia are usually composed of few well-identified organisms. Sometimes, those species do not co-inhabit the same environment, but their activities may be combined,163 Figure 13.
Phytoremediation with laccase gene
Phytoremediation for removal of recalcitrant environmental pollutants involve uptake, translocation, transformation and compartmentalization and sometimes mineralization (Figure 16).308 Plant roots also secrete metabolites, which stimulate the growth of micro-organisms in the rhizosphere, which in turn can degrade and mineralize the organic compounds. Although plants have several advantages over Fungi as candidates for bioremediation, they lack xenobiotic degradative capabilities unlike fungi. Hence, introduction of genes for degradation of recalcitrant environmental pollutants from microorganisms or other eukaryotes will further enhance their ability to degrade/mineralize recalcitrant environmental pollutants.
Microorganisms’ gene sequences are rich in CpG dinucleotides and have highly skewed codon usages, both of which are particularly unfavorable to efficient expression in plants.294 The MnP gene161 or the laccase gene328 from white- rot fungi has already been introduced into tobacco plants. Hirai et al.,151 reported laccase cDNA (scL) from white-rot fungus Schizophyllum commune was used as a model ligninolytic enzyme to obtain efficient expression of scL and removal of a recalcitrant compound in transgenic tobacco plant by decreasing the CpGdinucleotide motif content in order to develop phytoremediation with a ligninolytic enzyme-producing transgenic plant and a construct model in Figure 12.
Findings
Gene transfer into the fungi and myco-remediation
HGT and adaptation to different environments: Horizontal gene transfer (HGT), or lateral gene transfer, describes the transmission of genetic material between organisms, specifically across species boundaries.11,91,180,251 The term species is of course a difficult concept to apply to asexual microbes.71,90,189 HGT represents an important factor in shaping the genomes of prokaryotes and has provided a key source of evolutionary innovations.164,251 Several routes for transfer have been identified including gene transfer agents, transduction, transformation and conjugation.200,350 Fungi theoretically lack the beta-glucuronidase GUS) gene and therefore the metabolic capacity to utilize glucuronides as a carbon source. Wenzl et al.,385 screened for fungi in vertebrate urine using culture enrichment with selection for fungi with glucuronide metabolic capabilities. Using this approach they identified GUS genes in Penicillium canescens and Scopulariopsis spp. Subsequent, phylogenetic analysis demonstrated that these genes, along with GUS genes of Aspergillus and Gibberella, were derived by HGT from bacteria,350,385 again demonstrating cases of HGT which have expanded the metabolism of fungi and enabled them to adapt to new environments. The implication for the study Figure 10 shows the explosives fungal consortium isolates biodegradability in increasing order > Aspergillus niger >Trametes Versicolor > Rhizopus spp >Phanorochate chrysoporium > Penicillium spp. identified. Similarly to the Gunasekaran et al.,135 reported that, Rhizopus spp, Aspergillus niger, Trametes versicolor and P. chrysoporium as good mycoremediation potential for xenobiotic (Plate 1). The fungi-to-fungi transfers identified encompass five gene clusters representing a total of 53 individual gene phylogenies and seven gene cluster transfer events.184, 261,324,325,326 Perhaps the most striking example of a gene cluster transfer was the transfer of a 23-gene cluster from the Aspergillus lineage to Podospora.326 The majority of gene transfers between fungi (53 of 66 genes) were located in gene clusters, providing direct support for the hypothesis that gene clustering has played a role in facilitating gene transfer in fungi.380 Functional annotation of the 323 HGTs demonstrates that transfer has played a role in reconfiguring the core nutrient metabolism of many fungi, added to osmotrophic capacity of fungi, and allowed fungi to colonize ‘new’ environments. Mechanisms for gene transfer in fungi remain unclear, but the patterns of transfer (originating from bacteria and fungi) provide circumstantial evidence that supports two previously hypothesized routes of transfer: inter-domain transformation and conjugation-like transfer (for genes of bacterial origin) and anastomosis of cells (for those of fungal) reported.281
Surface sequestration
The fungi hyphal surface that is in direct contact with the environment consists of a cell wall, a cell surface component that is composed typically of chitin, 1, 3-β- and 1,6-β-glucan, and mannan along with different proteins and pigments (that is, melanin). Although, such interactions include physical adsorption, complexation, coordination, or chelation64,196 Figure 17, which do not depend on intracellular metabolism, their effects on the cellular metabolism remains unclear.111 The surface sequestration depends on three factors:
Biodegradation with extracellular enzymes with plethora of nonspecific extracellular enzymes are synthesized by fungi that can convert the complex extracellular growth substrates into simpler molecules, which can subsequently be absorbed into their cells as nutrients.378 The enzymes that are most beneficial Laccase gene transcription and biosynthesis, for example depends on a wide array of environmental cues, which include metals ions (e.g., copper (II) and iron (II)) and several aromatic compounds in the environment.111 In Figure 8, microbial fungi consortium (MFC) to 61.7% bioremediate heavy metal better than the isolated fungi species because the lateral gene transfer with where T. versicolor 359 in Aspergillus niger are HGT mediators and similar to Bohlin et al.,38 report.
Intracellular degradation
Fungal cells also possess the ability to catabolize internalized pollutants using their intracellular enzymes. One of the best studied enzyme systems used for this purpose comprises a set of mixed function oxidases called cytochrome P450s, which demonstrate the ability to catalyze a wide range of biodegradation reactions that involve epoxidation, hydroxylation, deamination, nitro reduction, dehalogenation, and desulfuration reactions.28,80,139, 341 Example of mycoremediation involving cytochrome P450 systems include biodegradation of kerosene and pharmaceutical waste products,63,131,213 dye decolorization 63 pesticide remediation.342,398 Mycoremediation reports the oxidoreductases that function hand-in-hand by Harms et al.,139 for pollutant degradation both within and outside of the cell (Figure 13). Most popular among these enzymes are the cytochrome P450 monooxygenase systems that are known to generate reactive oxygen species in eukaryotes274 and result in increased intracellular oxidative stress. Laccase is involved in the pigmentation process of fungal spores, the regeneration of tobacco protoplasts, as fungal virulence factors, and in lignification of cell walls and delignification during white rot of wood, 220 Figure 15 is an illustration of a construct intracellular degradation.
Kachia pollutants ecosystem restoration
Vegetation is not only an aid to ecosystem restoration, it is a key indicator of munitions xenobiotic.288 Plant species that are present on a site, as well as their quantities and condition, describe a watershed’s health and resilience. Kachia (Figure 19, Appendix 3) lies in the more southern Guinea savannah zone, Nigeria.140 Semi-arid zones of northern with soil fertility management strategies by smallholders that combine organic and inorganic inputs to sustain agricultural production.140Adopting best-fit legume technologies in cropping systems will contribute to improved soil fertility through biological N2-fixation and thereby increasing productivity of agricultural fields.112 Legume crop residue provides high quality fodder to feed livestock. Groundnut and soybean (Glycine max) variety TGx 1448-2E are widely grown by farmers are the common legumes and cultivate ginger for income generation and yam for food as well as income and the application of biofertilizer will help bio-remediate the residual pollutants within the ecosystems. Therefore, the use of the less efficient wild type tobacco plants could be combined with profit making operations,such as bioenergy cultivated (Figure 18) using PGPRs Biosurfactants presents in OTAI AG® Inocula in the biofertilizer (https://www.youtube.com/ watch? v=Hi_OpgVcFcg biofertilizer) from bio-waste in anaerobic digester inoculated with beneficial microbes that exhibit differing metabolic capabilities.255
Appendix 3
Kachia ecosystem restoration protocol is illuminated in Figure 15 where ‘Green liver model’ for the metabolism of xenobiotic in plants intracellular degradation role of fungi gene in planate and affirmed by scholar Burken et al.,55 Phytoremediation with protoplast fusion from micro-organisms, Figures 13 and 14. Most of the microorganisms’ enzyme activities could not be detected in microorganisms’ gene-introducing transgenic plants, and most studies in these transgenic plants have used Western blot analysis to detect these target proteins.179,241,234 Protoplasts have a negative charge on the surface that helps to repulse surrounding protoplasts. They need to be in close contact prior to fusion, which can be achieved physically (by mechanical pressing with micropipette, by centrifugation) or chemically. Protoplast fusion consists of three steps: agglutination, the fusion of membranes in small localized places and forming of bridges among protoplasts, rounding of fused protoplasts. Protoplast fusion requires approaching, adhesion and joining of two different types of protoplasts. Approaching of the protoplasts is determined by many electrostatic forces arising from the potential on the cell surface. The result of the interaction is either fusion or complete failure.33,37
Protoplast fusion
Somatic hybridization is an important tool of plant breeding and crop improvement by the production of inter-specific and inter-generic hybrids. The technique involves the fusion of protoplasts of two different genomes followed by the selection of desired somatic hybrid cells and regeneration of hybrid plants.104 Protoplast fusion provides an efficient mean of ‘gene transfer’ with desired trait from one species to another and has an increasing impact on crop improvement.53 Somatic hybrids were produced by fusion of protoplasts from rice and ditch reed using electro-fusion treatment for salt tolerance.237 In vitro fusion of protoplast opens a way of developing unique hybrid plants by overcoming the barriers of sexual incompatibility. The technique has been applicable in horticultural industry to create new hybrids with increased fruit yield and better resistance to diseases. Kachia pollutants ecosystem restoration for biodiversity of the shooting ranges is illustrated with Figure 16 xenobiotic myco /phytoremediation.
Green liver model
Phytoremediation is the term used to describe those methodologies that use living higher organisms, which include green vegetation, plants, aquatic plants, trees and grasses, to remove toxic compounds. This technology has the advantage of in situ treatment of contaminated soils, sediments, groundwater, surface water and external atmosphere. 81,217,351
To some extent, molecular biology approaches have already been used to evaluate phyto-remediation and reveal elimination of toxicity from contaminated sites.36,58,192,295 However, these sequences of microorganisms’ gene, which might be introduced into plants, are rich in CpG dinucleotides and have highly skewed codon usages, both of which are particularly unfavorable to efficient expression in plants.294 Practically, these MnP gene - or laccase gene-introducing transgenic plants have produced very low amounts of these enzymes.166,328 The mechanisms of degradation of xenobiotic by plants (Figure 14). Prior to the introduction of xenobiotic to plant cells, they must be taken-up through the roots. Research studies reports predictive relationships between the uptake rate of a compound and its physical-chemical properties.52,56 Enzymatic transformation of xenobiotic by plants follows the ‘green-liver model’ Figure 14 and involves three steps. First, the foreign compounds taken up by plants are transformed by enzymes such as cytochrome P450, carboxylesterases, and peroxidase.303 Secondly, the transformed xenobiotic are conjugated with D-glucose, glutathione, or amino acids188 by enzymes such as glutathione S-transferases, glucosyltransferases and malonyltransferases, resulting in either soluble or insoluble products. The third step is storage and compartmentation; the soluble compounds are stored in vacuoles or as cell-wall materials by further processing, and the insoluble compounds are generally assumed to be stored in the cell wall (Figure 14).209
The transformed products of TNT are further conjugated and sequestered in plant cells. Over 80% of the TNT label was associated with plant biomass, suggesting that the labelled carbon from TNT was sequestered in the plant tissues.138 The exemplar protocol for transformation and mineralization of TNT,156 RDX299 and HMX, in Figures 15 and 16 the xenobiotic biodegradation schematic. RDX biotransformation occurs in a variety of environments from surface and subsurface soils cold marine sediments410 and several microorganisms have been shown to co-metabolize both RDX and HMX.411 RDX and HMX has generally been thought to be recalcitrant to aerobic biodegradation411 and requires alternative biodegradation method like the Figure 15 biodegradations protocol affirmed by similar research report by Wang et al.,383 reported that, RDX is degraded via sequential reduction of the nitro functional groups followed by abioticring-cleavage.
Xenobiotic myco bio-augmentation/ phytoremediation/biosimulation (xenobiotic Myco B-P-B) techniques
The mechanisms by which TNT and its metabolites affect RDX and HMX degradation appear to lie in their cytotoxicity to most of the microorganisms present in the explosives - degrading culture reported.302 TNT and its degradation products are adsorbed to the upper soil layer, whereas RDX and HMX are not, being transported instead with the infiltrating water affirmed and reported302,402 that the potential for RDX biodegradation in the upper 4m is extremely high. The Kachia biodegradation and restoration of the military shooting range will requires xenobiotic myco /phytoremediation construct, Figure 16. Mycoremediation is a type of bioremediation using fungi, including WRF, for that purpose and it refers to their possibilities as microorganisms of degrading a great number of recalcitrant pollutants and transforming industrial wastes into products (Figure 16).194
Some of the most widely investigated fungi species capable of synthesising LiP are P. anerochaete chrysosporium, Trametes versicolor, Trichoderma reesei, Aspergillus niger, Phlebia radiata, Pleurotus ostreatus, Pleurotus sajor-caju.105 Biotransformation involving enzymes does not cause accumulation of toxic by-products, and the enzymes are digested after the completion of the process by the microorganisms dwelling in contaminated environments. Secondly, increasing the bioavailability of contaminants is more easily achieved than in the case of using whole cells.5,212 Laccase (benzenediol, oxygen oxidoreductases, EC 1.10.3.2) is one of the few lignin-degrading enzymes that have been extensively studied since 18th century. RDX degradation has also been examined in constructed wetlands.31,32 Both submerged and emergent plants were shown to take-up pollutants accumulated in sites of active growth. Biotransformation of RDX to unidentified products was also observed. Mineralization was very low (less than 5%) and production of volatile organics negligible. The nature of the chloromethane cycle137 transformation products remains unclear by Figure 16 xenobiotic protocol will optimize the environmental risk. Recently, laccases were reported from eukaryotes e.g. fungi, plants and insects.313 This makes laccases highly interesting for a wide variety of processes, such as textile dye decolouration, pulp bleaching, effluent detoxification, biosensors and bioremediation.78 Laccase is involved in the pigmentation process of fungal spores, the regeneration of tobacco protoplasts (Figure 13), as fungal virulence factors, and in lignification of cell walls and delignification during white rot of wood.220 Trametes versicolor is one of the most efficient laccase-producing fungi. Laccases are also produced in conjunction with LiP, MnP, or both. Laccases from marine fungi effectively mineralized 14C-(ring)-labelled synthetic lignin to14 CO2 .275 Laccase genes several strains of Trametes versicolor.169
Plants deal with organic explosives, such as RDX and TNT in three phases as depicted in Figure 14 and reported by scholars. 207,270,297 Phase I (transformation).The contaminant is metabolized into a more soluble and less toxic intermediate products by several reactions, such as oxidation, reduction, or hydrolysis. The oxidative metabolism of explosive compounds is generally mediated by cytochrome P450 mono-oxygenase in plants. In plants, cytochrome P450 forms a largest group of plant protein which plays an important role in degradation of explosives.236 Phase II (conjugation)-In conjugation between organic contaminant and endogenous hydrophilic molecules, such as D-glucose, glutathione, or amino acids, soluble or insoluble substances are produced to be subsequently sequestered in different cellular compartments of the plant for storage.307,406 Conjugation also enhances metabolic activity which is further catalysed by glycosyl-, malonyl-,and glutathione S-transferases. Phase III (compartmentation), (Figures 15&18).
Cytochrome P450-mediated phytoremediation
Cytochrome CYP superfamily of enzymes exist in all biological domains and their presence predates the emergence of oxygen-metabolizing life form.393 Cytochrome P450 (CYP) monooxygenases, the ubiquitous enzymes with catalytic versatility, substrate diversity and atypical kinetics are one of the most fascinating targets for biocatalysis and play diverse roles in biotechnology, medicine and bioremediation.51,120,195 Being a ubiquitous organism, fungi inhabitant’s diverse ecological niches, adapts to various sources of carbon and nitrogen for their survival and metabolism.130 Comprehensive biochemical analysis of molecular mechanisms showed that fungal adaption are often facilitated by CYP monooxygenases.130 Fungal CYPs play an essential role in their adaptations to ecological niches due to their diverse roles in the production of metabolites critical for pathogenesis, detoxification of xenobiotic and exploitation of substrates (Figure 2).130,265 To date, a vast number of CYPs have been identified in >2500 fungal species, and these are classified into ≈400 CYP families (namely CYP51-CYP69, CYP501-CYP699, and CYP5001-CYP6999).20,65
Under aerobic conditions, RDX is highly persistent in soil and groundwater; it is much less susceptible than TNT to biotransfonnation, is less strongly adsorbed to soil, and undergoes much less immobilization.321 Some notable examples of the fungal primary metabolism are housekeeping functions such as ergosterol biosynthesis, meiotic spore-wall biogenesis, and n-alkane hydroxylation, whereas fungal secondary metabolism deals with the biosynthesis of hormones, mycotoxins, and the like (Figure 15) reported by Črešnar and Petrič.80 Fungal CYPs are also capable of detoxifying and degrading various xenobiotic compounds encountered in their environments,345,346 such as polycyclic aromatic hydrocarbons (PAHs), phenolic compounds, and other toxic environmental pollutants.80 Remarkably, the transcriptional regulation of CYP- dependent metabolic pathways can be alternatively induced or up-regulated by defined high or low nitrogen conditions, as observed in P. chrysosporium, C. versicolor, P. placenta, and A. oryzae (Figure 17).170,219,242
Considering the extraordinary potential of CYPs, several reviews have addressed their limitations and have focused on tackling these challenges to promote CYPs as robust (Figure 15) biocatalysts.29,195,224,250 Fungal CYPs also inherently possess the same limitations; and recent advancements has sought to overcome the major hurdles posed by fungal CYPs.29,61 Enzyme engineering through mutagenesis is one of the tools to modify fungal CYPs as sustainable catalysts by solving issues concerning low expression levels and poor activity.29,195,224,250 For example, the oxidizing activity of the P. chrysosprium PAH oxidase (CYP5136A3) was significantly improved by a rational designing approach through site-directed mutagenesis.224 Artificial CYP-CPR fusion constructs termed “Molecular lego” are a competent strategy for comparative analysis of differential redox partners to find the optimal redox system (Figure 16).300,301 In addition, improved intracellular electron recycling can be obtained through cofactor regeneration369 by co-expressing CYPs along with glucose/formate dehydrogenase or aldehyde reductase (Figure 15).206
Biofertilizer-soil amendments
The use of soil amendments such as compost,396 biochar, biofertilizer and molasses to ‘kick-start’ microbial degradation has been well studied for many xenobiotic reported by Wu et al.397 and Otaiku et al.,255 But few studies have yet investigated the effect of amendments for phytoremediation on explosives. A laboratory study199 showed that the application of molasses to open burn/detonation areas enabled Guinea Grass (Panicum maximum) to remediate RDX, with plant and microbial communities’ fully remediating soil contaminated with 1.3 mg/kg RDX within the 15-week study. When biofertilizer applied as soil inoculants, they multiply and participate in nutrient cycling and benefit crop productivity.319
Depending on the family of N.tabacum 30-40% of the seed is in average oil129,360 and is composed of linoleic acid (71.63%), oleic acid (13.46%) and palmitic acid (8.72%) . Although tobacco seed oil is a non- edible vegetable oil, it can be utilised for biodiesel production as a new renewable alternative diesel engine fuel.364, 373 For this purpose research has to focus on the amounts of inorganic and organic pollutants accumulated in the seeds during the phytoremediation process.
Tobacco is a plant, which fulfils all the characteristics for a suitable phytoremediation plant and combined with the production of bioenergy it could become one of the main plants used in the field of phytoremediation. Tobacco is a plant, which fulfils all the characteristics for a suitable phytoremediation plant and combined with the production of bioenergy. Niche fungi consortium research on diversity and distributions59,103,183,365,387 will enhance the re-construction ofthe Kachia biogeography spatial scale relevant to ecology and evolution.141 The convergence of biogeography and ecology is a relatively recent phenomenon282,285,286 species sorting and niche partitioning; local/regional saturation; and diversification of regional biotas)284 analyzes organism-environment relations through change over space and time, and often includes human-biota interactions (Figure 13).
Site monitoring
The significant cost factor of site remediation is monitoring of contaminant levels, and this is particularly difficult given the large scale, and inaccessibility of military sites. Novel studies have quantified the response to explosives of plant species native to military ranges in temperate regions. By measuring multiple variables representing morphological and physiological responses, such studies identified vegetation responses that could potentially be used to determine levels of TNT and RDX contamination.7,374,375 Hyper spectral imaging technologies are also being developed to remotely detect vegetation response using aerial drones as suitable carriers to record contamination levels on surface in accessible sites.
Myco B-P-B techniques–examplar Despite several studies, bioremediation of TNT is still considered challenging. Majority of studies have focused on discovering microorganisms that could be used to degrade pollutants was reported by Ayoub et al.,15 and No˜lvak et al.,247 research reported the effect of bioremediation methods and combinations (biostimulation, bioaugmentation, phytoremediation) on the degradation of TNT in soil. Biostimulation approaches add nutrients or electron acceptor/donors to the polluted environment to enhance the bioremediation of pollutants like TNT.125 Biostimulation can also be used in combination with bio-augmentation to improve the survival and catabolic activity of introduced microorganisms.209 Phytoremediation
Phytoremediation utilizes green plants to treat contaminated soil or water and has been successfully applied to remediation of different pollutants, including TNT.153,208 Plants release root exudates and enzymes that stimulate microbial activity and contaminant degradation in the rhizosphere, enabling the application of rhizoremediation, which is considered particularly effective for the treatment of contaminated soil (Figure 16).124,193,308 Using 16S rRNA gene full-length sequencing (BCCM/LMG, Belgium), the predominant bacterial strains in the inoculum were determined to be members of Pseudomonas and Stenotrophomonas genus.200
The research indicates that all treatments used resulted in decreased concentrations of TNT in soil. Similar to previous reports.354,355, 370 TNT and its aminoderivates 2ADNT and 4ADNT were the only compounds detectable by HPLC; all of these were reduced to a considerable degree of the initial concentrations. The isomeric aminoderivates (2ADNT and 4ADNT) are formed during microbial transformation of TNT;395 these compounds exhibit less toxicity than TNT and bind more tightly to soil clay particles and organic matter, thereby reducing bioavailability as well as toxicity in soil.264,273Of the treatments used, the combined bioaugmentation - biostimulation approach coupled with plant (especially rye) cultivation was the most effective treatment resulting in the lowest final TNT concentrations. This corresponds well to other more recent results with similar TNT contamination rates.355
Future work
The Figures 14–16 xenobiotic myco /phytoremediation construct are an integrated approach to metabolic engineering has beentermed inverse metabolic engineering, where first a desired phenotype is formulated, then the necessary factors to create it are determined, finally these are assembled in the host (microbial/Phyto) as affirmed by Bailey et al.,16 as a construct for xenobiotic biodegradation model called PC3R technology. Finally, incremental genetic changes in vivo and input the results of each change back into the model. The technological and methodological tools now exist to accomplish PC3R technology where Aspergillus as exemplar as gene mediator. Further work on PC3R technology with respect to xenobiotic biodegradation will require the solution mathematical modelling of metabolic pathway networks; the theory is well developed at present244,389 and comprises several complementary approaches to metabolic modelling, e.g., stoichiometric and carbon flux models (metabolic flux analysis339,371 kinetic models (metabolic control analysis and morphologically structured models. 233,245,323 The development of remediation approaches is the determination of the genomes of specific white rot fungi. For example, research into the P. chrysosporium genome has resulted in the elucidation of specific cytochrome P450 monooxygenases, which may be differentially expressed in the presence of xenobiotic compounds.400
In the future, efforts should be made to develop strategies to improve the tolerance, uptake, and yperaccumulation of heavy metals/metalloids using genomic and metabolic engineering approaches. Pathways that control the uptake, detoxification, transport from root to shoot tissues and translocation and hyper-accumulation in the aboveground storage tissues can be engineered using gene-stacking approaches. Genome editing strategies can be designed using TALENs (transcription activator like effectors nucleases) technology or the powerful CRISPR- Cas9 (clustered regularly interspaced short palindromic repeats) system to produce microbes/plants for bio/phytoremediation purposes (Figures 17&18). Recently, efficient and successful CRISPR/Cas9- mediated targeted mutagenesis has been reported in Populus plants.106
There is a need for the development of ex-situ bio-augmentation products for the biodegradation of xenobiotic to assist mineralization because the additional nitrogen supports microbial growth without competing with the explosive compounds for metabolism of RDX as reported by scholars291,329,379 and TNT41 nitrogen-bearing compounds are also potential inhibitors of enzyme expression, as presented by Nejidat et al.,243 who showed that ammonium and nitrite repress fungi lignin peroxidase. Extracellular oxidoreductases. A unique characteristic of fungi is their ability to attack organic compounds using a range of extracellular oxidoreductases with relatively nonspecific activities. These enzymes have probably evolved to support fungal growth on recalcitrant substrates of random structure such as lignocellulose substrates that are not accessible for most bacteria through the depolymerisation and removal of lignin and its humic substance derivatives.18,154,269 Hence, these enzymes give fungi an ecological advantage over bacteria in particular decomposition niches.18 Genetic engineering of peroxidases to improve their catalytic properties allows enhancement of the H2O2 resistance of the enzymes, or the structural modification of already commercially available enzymes to enable the oxidation of other compounds with high redox potentials.296 Radical reactions may also covalently bind metabolites (for example, of TNT) to soil organic matter.144,248 Cytochrome P450 monooxygenases95 also contribute to the catabolic versatility of fungi lacking extracellular oxidoreductases.
None.
The authors declare that there was no conflict of interest.
Authors had no funding for this research.

©2020 Otaiku, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.