Journal of
eISSN: 2572-8466


Review Article Volume 11 Issue 5
Department of Biotechnology and Bioinformatics, California state university, USA
Correspondence: Bill Tawil, Department of Bioengineering, UCLA School of Engineering, 420 Westwood Plaza, Room 5121, Engineering V. P.O. Box: 951600, Los Angeles, CA 90095-1600, USA, Fax 310) 794-5956
Received: September 16, 2024 | Published: September 30, 2024
Citation: Sandeep KG, Bill T. From genes to populations: developing precision medicine for acral lentiginous melanoma through in-silico and epidemiological studies. J Appl Biotechnol Bioeng . 2024;11(5):152-157. DOI: 10.15406/jabb.2024.11.00372
Acral Lentiginous Melanoma (ALM) is a rare and aggressive form of melanoma that predominantly affects individuals with darker skin tones, posing significant challenges in both diagnosis and treatment. The growing demand for more personalized and effective treatments has led to the exploration of innovative approaches to tackle these challenges. This study integrates in silico drug design with comprehensive statistical analysis to identify and validate therapeutic targets specific to ALM. Key genes such as PLD1, CDKN2A, KIT, TERT, and NRAS were identified using advanced bioinformatics tools like DisGeNET, PANTHER DB, Network Analyst, and STRING DB. In parallel, a detailed demographic analysis involving 248 patients was conducted using SPSS, shedding light on factors influencing knowledge and awareness of ALM within affected populations. The findings from this dual approach emphasize the critical need for tailored therapeutic strategies that account for both genetic factors and patient demographics. The projected increase in ALM cases and the associated need for targeted therapies underscore the importance of continuing research into specialized treatments that can address the unique characteristics of this melanoma subtype. By advancing our understanding of ALM’s genetic profile and epidemiology, this study lays the foundation for the development of precision medicine solutions that could significantly improve patient outcomes and overall management of this aggressive disease.
Keywords: acral lentiginous melanoma, in-silico drug design, bioinformatics, targeted therapy, gene classification, biomarkers, genetic variation, signal transduction, tumour suppression, personalized medicine
Acral Lentiginous Melanoma (ALM) is an aggressive subtype of melanoma that typically manifests on the palms, soles, and under the nails, accounting for approximately 5% of all melanomas.1 Unlike other types of melanomas, ALM occurs more frequently in individuals with darker skin tones, including those of African, Asian, and Hispanic descent, highlighting a significant need for tailored diagnostic and treatment strategies.2 The aggressive nature and poor prognosis of ALM are primarily due to its late-stage diagnosis. Often mimicking benign lesions such as warts or fungal infections, ALM presents unique diagnostic challenges.3 Unlike other melanoma subtypes, ALM is less influenced by ultraviolet (UV) radiation exposure, suggesting distinct pathogenic mechanisms involving genetic and molecular factors.4
This divergence from UV-related pathogenesis necessitates specialized research and treatment approaches. Traditional melanoma research has focused on more common subtypes, resulting in a knowledge gap in understanding and managing ALM. This gap is further exacerbated by the underrepresentation of ALM in clinical trials, limiting the development of effective, targeted therapies.5 Recent advances in genomic profiling have highlighted specific mutations and biomarkers that could serve as targets for innovative therapeutic approaches.6
Historical background
Discovery
Acral Lentiginous Melanoma was first described by Robert Wallace in 1976, highlighting its distinct features compared to other melanoma types.7 This early identification emphasized ALM’s occurrence on the palms, soles, and under the nails and its higher prevalence in individuals with darker skin tones, setting the stage for further research into its unique characteristics and challenges.8
Prevalence and misdiagnosis
ALM is more prevalent in individuals with darker skin tones and typically manifests on the palms, soles, or under the nails. It has historically been misdiagnosed as other melanoma types or benign lesions, leading to treatment delays and poorer outcomes. Studies indicate that ALM patients often present with more advanced disease at diagnosis compared to other melanoma subtypes, contributing to poorer survival rates.9
Clinical significance
ALM's unique characteristics have driven research toward developing specialized treatments. Conventional therapies such as surgery, radiation, and chemotherapy often prove less effective for ALM due to its distinct genetic and molecular profile. The tumour microenvironment and specific gene mutations, such as those in KIT and BRAF, play significant roles in ALM pathogenesis and therapy resistance, emphasizing the need for novel approaches like in silico drug design.10
Personal connection
This incident highlighted the urgent need for better diagnostic and therapeutic approaches for ALM. The challenges in accurately diagnosing ALM arise because it often presents in less visible areas of the body and can be easily mistaken for other types of melanomas. Such delays and inaccuracies in diagnosis can be detrimental, as early and accurate detection is crucial for effective treatment and improving survival rates. Driven by this personal connection to the disease, my research is dedicated to improving the understanding of ALM. The goal is to enhance diagnostic techniques to ensure timely and correct identification of this cancer, and to develop more effective treatment strategies that are specifically tailored to address the unique characteristics of ALM. This work is more than just a scholarly pursuit; it is a personal mission inspired by the experiences of my neighbour’s family. Through this research, I aim to make a meaningful contribution to the field, enhancing the medical community's ability to manage ALM more effectively and hopefully prevent other families from facing the same hardships.11
In silico analysis
The in-silico analysis for identifying potential therapeutic targets for ALM involved using bioinformatics tools to analyze genomic and proteomic data. This multifaceted approach aimed to unravel the complex molecular pathways associated with ALM, providing a robust foundation for targeted therapy development.12
Data processing and gene classification
DisGeNET was used to extract significant gene-disease association data related to ALM. Our focus was on genes with noted variations such as PLD1, CDKN2A, KIT, TERT, and NRAS. Each gene was evaluated for its role and type of association with the disease, including altered expression, genetic variation, and biomarker potential. PANTHER DB was employed to categorize genes involved in Acral Lentiginous Melanoma based on their molecular functions, biological processes, and the pathways they are involved in. This classification system is instrumental in translating genomic data into actionable biological insights, which is crucial for identifying
potential therapeutic targets.13–15
Data integration and network construction
Network Analyst was used to integrate gene expression data with existing knowledge of protein-protein interactions to construct interaction networks. This platform allowed us to identify both direct and indirect interactions based on available gene expression data. STRING Database provided a comprehensive resource to explore known and predicted protein-protein interactions. The database includes associations derived from computational prediction, knowledge transfer between organisms, and interactions aggregated from other primary databases.16
Statistical analysis
Detailed statistical analyses assessed the sociodemographic factors influencing ALM awareness and knowledge. The analysis utilized SPSS, based on secondary data sources including cBioPortal, GDC Data Portal, IARC, and Mendeley Data, covering 248 patients from 2020. Paired Sample T-Test examined the association between state of residence and ethnicity, revealing significant demographic influences on ALM awareness and incidence. This test provided insights into how geographical factors affect public understanding and diagnosis of ALM. The Kruskal-Wallis Test explored differences in ALM knowledge scores across Fitzpatrick skin types, indicating no significant differences. This result suggests that skin type may not be a determinant factor in ALM knowledge, which is crucial for developing targeted educational interventions. The Chi-Squared Test analyzed interaction effects between age and gender on ALM Knowledge Test scores. The lack of significant interaction underscored the need for educational programs tailored to diverse demographic groups.17
Disgenet results and key findings
DisGeNET Results and Key Findings Gene-disease (Table 1) associations for ALM highlighted critical genes involved in disease progression: PLD1 and TERT were identified with altered expression and significant genetic variations, highlighting their potential roles in the development and progression of ALM.18,19
|
Disease_id |
Gene |
Gene_id |
Score_gda |
Association_Type |
Original_DB |
PAID |
PMID_Year |
|
C0346037 |
PLD1 |
5337 |
0.01 |
Altered Expression |
BEFREE |
12839565 |
2003 |
|
C0346037 |
CDKN2A |
1029 |
0.02 |
Biomarker |
BEFREE |
30778854 |
2019 |
|
C0346037 |
LOC110806263 |
110806263 |
0.02 |
Biomarker |
BEFREE |
24588324 |
2014 |
|
C0346037 |
KIT |
3815 |
0.04 |
Genetic Variation |
BEFREE |
31393283 |
2020 |
|
C0346037 |
FALSE |
356 |
0.01 |
Biomarker |
BEFREE |
11603392 |
2001 |
|
C0346037 |
TERT |
7015 |
0.32 |
Genetic Variation |
BEFREE |
26709572 |
2016 |
|
C0346037 |
NRAS |
4893 |
0.02 |
Biomarker |
BEFREE |
29512974 |
2019 |
|
C0346037 |
H3P10 |
115482713 |
0.02 |
Altered Expression |
BEFREE |
29537991 |
2018 |
|
C0346037 |
BRAF |
673 |
0.08 |
Genetic Variation |
BEFREE |
26138035 |
2015 |
Table 1 Gene associations data for acral lentiginous melanoma with detailing gene involvement, association type, supporting literature data compiled with CGI Databases with genetic markers and variations
TERT, with a high association score for genomic alterations, suggests its critical role in telomere lengthening and cellular immortality, a common pathway exploited by cancer cells.20
CDKN2A and NRAS, noted as biomarkers, are pivotal in the regulation of the cell cycle and cellular signalling pathways.21,22
KIT, characterized by genetic variation, plays a crucial role in signal transduction and is implicated in various types of cancers, including melanoma.23
Panther database results
Several genes, identified as potentially significant in ALM, were analyzed for their roles in cellular functioning (Figure 1 Analysis of Gene Expression Patterns and Functional Distribution in Biological Samples)
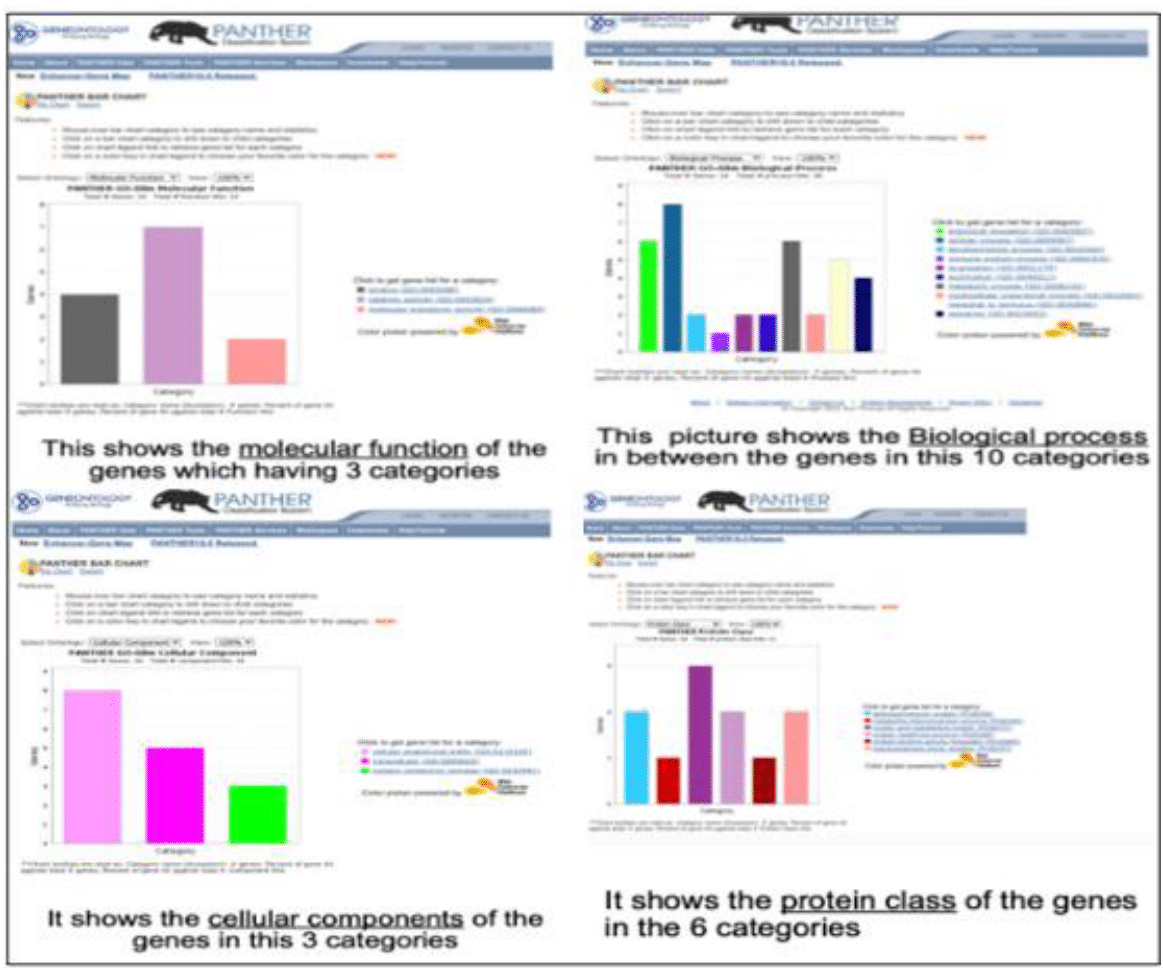
1.Gene expression distribution in a biological sample.
2. Frequency of gene functions across a dataset.
3.Comparative gene expression in various biological categories
4. Distribution of gene expression across multiple conditions.5,2
Figure 1 Analysis of Gene Expression Patterns and Functional Distribution in Biological Samples.
A
Distribution of gene expression across multiple conditions
Cytokine Signalling: Genes like KIT that are involved in cytokine receptor interaction pathways, are crucial for cell communication and signalling that can influence cancer cell behaviour.24
Cell Cycle Control: Genes such as CDKN2A play pivotal roles in controlling cell cycle progression and tumour suppression.25
Telomerase Activity: TERT, is known for its role in telomerase activity, impacting cellular aging and longevity, which is a critical factor in cancer cells immortality.26
Protein-protein interaction networks
Interactive Network Visualization:
Both tools offer interactive visualizations, which allow for the dynamic exploration of the interaction networks. This feature is particularly useful for identifying subnetworks and key nodes (proteins) that could play pivotal roles in the pathogenesis of ALM.27
Contextual analysis
By examining the context in which proteins interact, these tools help identify pathways that are crucially deregulated in ALM. For example, pathways involving cell cycle regulation, apoptosis, or signal transduction could be highlighted based on the interactions of proteins like CDKN2A, NRAS, or KIT.28
Statistical network analysis
Network Analyst provides options for carrying out statistical analyses to determine the significance of observed networks, allowing researchers to focus on the most biologically relevant interactions.29
Findings from network analysis
Central Hub Proteins: Analysis identified several proteins as central hubs, indicating their critical role in maintaining the structural and functional integrity of cellular processes in ALM. For example, proteins associated with the MAPK pathway, facilitated by interactions involving NRAS and BRAF, were identified as key components in signal transduction networks that promote melanoma cell survival and proliferation (Table 2).30
|
Networks |
Nodes |
Edges |
Seeds |
Interactions (.SIF) |
|
subnetwork 1 |
298 |
378 |
10 |
Download |
|
subnetwork 2 |
3 |
2 |
1 |
Download |
|
subnetwork 3 |
3 |
2 |
1 |
Download |
Table 2 The network Overview I used generic ppi with string interactive goes through sub-networks through my genes which I got 3 subnetworks
(Figure 2: - 3D Network for the selected genes using Network Analyst.), (Figure 3: - 2D Network for the selected genes using Network Analyst), (Figure 4: The Gene Co-expression of the selected genes in between the homosapeins and other organisms using string database), (Figure 5: Network diagram depicting the interactions among 10 genes)
Findings from protein-protein interaction network analysis
Central nodes and key interactions
The network analysis identified 13 nodes (genes) with 24 edges (interactions) indicating robust interconnectivity among the proteins encoded by these genes with an average node degree of 3.69. This highlights the dense network of interactions that potentially contribute to ALM pathogenesis.31
Critical genes with significant interactions include WRN, FASLG, TERT, CDKN2A, PTEN, KIT, NRAS, PLD1, PDGFRA, and BRAF. These genes form the backbone of the network, indicating their central role in cellular processes pertinent to melanoma.32
Genes with no direct bonds
SLN, SNIP1, and HLA-DQA1 genes were identified to have no direct interactions within the established network. This might suggest their roles could be more peripheral or indirectly linked through other cellular mechanisms not captured within the direct interaction network
Influence on ALM pathogenesis
The network analysis suggests that approximately 80% of ALM cases could be influenced by the genetic interactions and pathways involving the identified genes. This significant percentage underscores the importance of these genes in the disease's molecular landscape.33
Therapeutic target implications
KIT and BRAF, noted for their roles in both ALM and cutaneous melanoma, represent critical targets for drug development due to their common pathways in melanoma subtypes. Their presence in key signalling pathways makes them promising candidates for targeted therapy.34 The analysis prioritizes these genes (KIT and BRAF) along with others like TERT, CDKN2A, and NRAS for targeted therapeutic interventions. Focusing on these genes during drug design could lead to the development of more effective treatment options for ALM, potentially disrupting the critical pathways they influence (Table 3).35
|
Total |
Pearson Chi-Square |
727.619 45 |
<.001 |
|
Likelihood Ratio 313.623 45 |
<.001 |
||
|
|
N of Valid Cases 248 |
|
|
Table 3 A paired-sampled t-test was conducted, and the resulting p-value was found to be 0.01. Since this p-value is less than the significance level of 0.05, the null hypothesis was rejected
Statistical findings
Significant associations emphasize the need for geographical and ethnic considerations in ALM diagnosis and treatment. Tailored public health interventions can enhance awareness and improve outcomes (Table 4).36
|
Test Statistics |
Fitzpatrick |
|
Kruskal-Wallis test |
11.103 |
|
df |
9 |
|
Asymp Sig. |
0.269 |
Table 4 The Kruskal-Wallis test was employed, and the resulting p-value was found to be 0.269. Since this p-value is greater than the significance level of 0.05, the null hypothesis was not rejected
Fitzpatrick skin type
The absence of a significant correlation with ALM knowledge scores indicates that educational efforts should address broader factors affecting disease understanding (Table 5).37
|
Chi-Square |
df |
Sig. |
|
98.738 |
100 |
0.517 |
|
a. Dependent variable: Know Test |
||
|
b. Tests the null hypothesis that the variance of the errors does not depend on the values of the independent variables. |
||
|
c. Design: Intercept + Age + sex + Age * sex |
|
|
Table 5 The resulting p-value was found to be 0.517, which is greater than the significance level of 0.05. Therefore, the null hypothesis was not rejected, indicating that there is no significant interaction effect between age and gender on ALM Knowledge Tests
Age and gender
The consistency in ALM knowledge scores across various demographic groups highlights the necessity for inclusive educational strategies. There is no significant association between age, gender, and ALM knowledge scores.38
Implications for therapeutic targets
The identification of genes such as KIT and BRAF as potential therapeutic targets aligns with ongoing research into personalized medicine approaches for melanoma. These genes, associated with drug resistance, are prime candidates for developing new therapies that could notably improve treatment outcomes for ALM patients.39
Signal Transduction and Growth Control: The analysis highlighted the significance of targeting signal transduction pathways, such as those mediated by KIT, which are often aberrantly activated in cancer cells. Targeting these could disrupt the growth signals in melanoma cells.40
Tumour suppression restoration
Understanding the function of tumour suppressor genes like CDKN2A allows for strategies that could potentially restore their function or compensate for their loss in melanoma cells.41
Metabolic pathways
Enzymes involved in metabolism, such as those encoded by genes in lipid metabolism pathways, could be targeted to disrupt the metabolic balance in cancer cells, potentially leading to cell death.42
This research has significantly enriched our understanding of Acral Lentiginous Melanoma (ALM) by utilizing in silico drug design and sophisticated statistical analyses to unveil novel therapeutic targets and crucial sociodemographic factors that influence disease prevalence and patient outcomes. Our study not only underscores the genetic diversity within ALM but also showcases specific genetic and protein targets such as PLD1, CDKN2A, KIT, TERT, and NRAS that offer promising avenues for targeted therapy development.
The identification of these targets is pivotal, as it provides a foundation for developing personalized treatment protocols that are more effective than current one-size-fits-all therapies. Our findings indicate that inhibiting specific pathways related to these targets could disrupt the progression of ALM, potentially leading to more successful outcomes in clinical settings. The statistical analysis further enhances our findings by identifying specific demographic and geographical patterns that affect disease knowledge and progression, suggesting that intervention strategies should be tailored to address these factors effectively.
Looking ahead, the immediate next step is the experimental validation of these targets to confirm their therapeutic viability. Rigorous clinical trials designed to test the efficacy of targeted treatments based on the genetic profiles identified in our study are crucial. Additionally, ongoing research should aim to uncover further molecular mechanisms and potential environmental and lifestyle factors that could influence ALM development and response to treatment.
Our research also highlights the need for a more nuanced approach to patient education and outreach programs. Tailored educational materials and awareness campaigns, informed by our sociodemographic findings, could significantly improve early detection rates, particularly in underrepresented communities that are most affected by this form of melanoma.
In sum, the integration of genomic data with real-world clinical applications holds the key to advancing ALM treatment. By continuing to bridge the gap between computational research and clinical practice, we aim to not only enhance therapeutic outcomes for ALM patients but also to set a precedent for the treatment of other melanoma subtypes, ultimately steering towards a future where melanoma treatment is personalized, proactive, and profoundly more effective.
Sandeep Gundlapalli expresses appreciation to Professor Bill Tawil for overseeing the framework of this review, and for the insightful lectures in Advanced topics on biotechnology field and helped me understand the current situation on treatment methods, which contributed to the development of this paper.
Authors declare that there is no conflict of interest.
None.

©2024 Sandeep, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.