Journal of
eISSN: 2572-8466


Research Article Volume 5 Issue 2
University School of Biotechnology, Guru Gobind Singh Indraprastha University, India
Correspondence: Prakash Chand Sharma, University School of Biotechnology, Guru Gobind Singh Indraprastha University New Delhi 110078, India, Tel 91-11-25302306, 25302123, Fax 91-11-25302111
Received: February 22, 2018 | Published: March 30, 2018
Citation: Rai PS, Chaudhary S, Sharma PC. Expressed sequence tags (ESTs) – based computational identification of novel and conserved microRNAs in turmeric ( Curcuma longa L.). J Appl Biotechnol Bioeng. 2018;5(2):112-119. DOI: 10.15406/jabb.2018.05.00125
MicroRNAs (miRNAs) are endogenous, small, single stranded non‒coding 17‒24 nucleotide long RNAs that regulates gene expression in plants and animals either by direct degradation of mRNA or inhibition of translation. Conventional approaches employed for the detection of miRNA are costly and time consuming. On the other hand, comparative genomics assisted with modern tools of bioinformatics provide an efficient and cost‒effective identification of novel and conserved miRNAs via homology search within known miRNAs. The present study reports computational identification of miRNAs present in the Expressed Sequence Tags (ESTs) of turmeric (Curcuma longa L.), a plant known for its great medicinal and culinary value. A total of 12678 ESTs were assembled into 2710 contigs/unigenes and homology sequence search was performed against 4647 non‒redundant miRNAs of Viridiplantae. In total, 102 potential mature miRNAs showed homology with 51 contigs/unigenes. Subsequently, four novel miRNAs were identified in turmeric, which satisfied the potential miRNA criteria. These four miRNAs were further characterized for target gene prediction identifying 10 target genes with their putative functions. Our study provides a valuable resource on miRNAs in turmeric, and also suggests a methodology useful for the identification of novel and conserved miRNA in non‒model but important organisms, whose EST and genome sequence data are available in the public domain.
Keywords: turmeric;, Curcuma longa, microRNA, expressed sequence tags
miRNA, MicroRNA; EST, expressed sequence tags; cDNA’s, complementary DNAs
The discovery of novel regulatory RNAs opened a new horizon in molecular biology research. Till recent past, protein coding genetic constituents have been used to detect different regulatory mechanism of gene expression. However, protein coding genes constitute a very small portion of the whole genome suggesting that the genome is not all about protein coding regions. MicroRNAs (miRNAs) are small, single‒stranded, endogenous and non‒coding ~22 nt RNAs, present in plants, animals and some of DNA viruses, where they play important regulatory roles in gene expression by mRNA cleavage or repression of translation.1‒3 Known 1881 sequences of miRNAs account for >3% of all human genes.4 Phylogenetically, miRNAs are highly conserved across species.2,5
MiRNAs are transcribed as primary‒ transcript (pri‒miRNA) from different genomic locations by RNA polymerase ІІ.6 They are present in either non‒coding RNAs or the introns of protein coding genes.7 Subsequent to transcription, Drosha and Dicer, two members of RNAase‒ІІІ enzyme family, process pre‒miRNA into precursor miRNA (pre‒miRNA). This pre‒miRNA is ~70‒200 nt long that folds into a stem and loop structure with multiple bulges and mismatches. The pre‒miRNA is exported out of nucleus into cytoplasm by the Exportin‒5 (Exp‒5), in a Ran‒GTP dependent manner. The pre‒miRNA, in the cytoplasm, is processed and cleaved by Dicer to generate a ~20‒bp duplex intermediate.8 Only one strand of the duplex compiles mature miRNA as per thermodynamic symmetry rule.9 Subsequently, mature miRNA gets assembled into the effector complexes known as miRNPs (miRNA‒containing ribonucleoprotein particles).7 On the basis of complementarity, miRNP guides its binding with target mRNA. Mechanism of target recognition by miRNAs is different in plants and animals. In plants, miRNAs bind to a generally perfect, single complementary site in either coding or 3´ un‒translated regions (3´‒UTRs) of the target mRNA, whereas, in animals, the miRNAs bind to multiple and partially complementary sites in the 3´‒UTRs. Additionally, coding or 5´‒UTRs targets are also functional.10 Depending upon the base pairing to miRNA, mRNA may be directly destructed on perfect or near‒perfect complementarity.11 Alternatively, miRNA will inhibit translation causing inhibition of protein accumulation.2 The flow chart representation of biogenesis and function of miRNA in eukaryotes is provided in the Figure 1.
The Expressed Sequence Tags (ESTs), single stranded subsequence of complementary DNAs (cDNAs), were first generated during Human Genome Project.12,13 Traditional genomics approaches are limited to model organisms but EST sequencing being labour and cost effective provides a good platform for studies on structural and functional genomics for even non‒model organisms. ESTs are more important for eukaryotes since they have large genome size and low gene density. Hence, ESTs provide an efficient resource in the context that they have very high functional information content and also often correspond to known or predicted/hypothetical function.14 The computational analysis of expressed sequence tags for miRNA identification has been extensively exploited in different plant species, including those with high medicinal, ecological, and nutritional value, such as jatropa,15 garlic,16 beet17 and many more. In the present study, EST based approach has been exploited to identify novel and conserved miRNAs in turmeric, a plant with immense medicinal and economic importance.
Turmeric (Curcuma longa L.), a member of the family Zingiberaceae (ginger family) is an endemic herb of south‒west India and now used and cultivated in many parts of Asia including almost all parts of India. Curcuminoides, a group of compounds including curcumin (di‒ferulolymethane), demethoxycurcumin and bis‒demethoxycurcumin, are the key chemical components of the turmeric. Of these, curcumin, present in abundance in turmeric powder, is responsible for most of the medicinal and economical value of turmeric. The other important constituents of turmeric powder include turmerone, atlantone, and zingiberene.18 Turmeric has diverse uses viz. culinary, medicinal, dye, etc.19 Being such a valuable plant species, medicinally as well as economically, turmeric has attracted the attention of many molecular biologists worldwide.
In the present study, we have identified conserved and novel miRNAs in the Expressed Sequence Tags (ESTs) of turmeric available in the public domain.
Computational resources
A total of 8439 mature miRNAs of Viridiplantae were downloaded from miRBase Release 21 (http://www.mirbase.org/cgi‒bin/browse.pl), a database of miRNA sequences and annotation.20 To identify conserved and novel miRNAs in turmeric, 4647 unique and non‒ redundant miRNAs were selected for further analysis. A total of 12678 EST sequences of turmeric (Curcuma longa L.), were downloaded from the dbEST database of NCBI (http://www.ncbi.nlm.nih.gov/nucest/). Offline Blast version, Blast 2.2.30+was downloaded from the NCBI website (ftp://ftp.ncbi.nlm.nih.gov/blast).
EST assembly and functional annotation
CLC Genomics Workbench 8.0 (http://www.clcbio.com/products/clc‒genomics‒workbench/) was used to assemble expressed sequence tags (ESTs) downloaded from NCBI. The assembled ESTs were further analyzed using, BLAST2GO suite21 for functional annotation and assigning gene ontology (GO) term. The similarity search was performed against the existing annotated sequences present in public database using BLASTn and BLASTx module followed by mapping and annotation. To keep the stringency level high during annotation of unigenes, the entire similarity search was performed by keeping E‒value at 10.
Identification of mature miRNAs in assembled ESTs
A local database of mature miRNAs, downloaded from miRBase, was made using BLAST tool. The algorithm of BLAST was used for the pair wise alignment of the assembled EST sequences, as query against unique miRNA database keeping threshold of E‒value at 10. The filter was used to choose low complexity, word size of 7 and window size of 7 between query and database.
For the selection of candidate miRNAs from the homology search, threshold was set at 18 nt length with no gap in between.16 The assembled sequences which matched closely to the known miRNAs were selected for further study. The mismatch was taken as less than three nucleotides. To remove the protein coding region, hits obtained after satisfying earlier mentioned criteria were further subjected to BLASTx search against‒non redundant (NR) protein database. The diagrammatic representation of the scheme identification of miRNA in turmeric and EST analysis is shown in Figure 2.
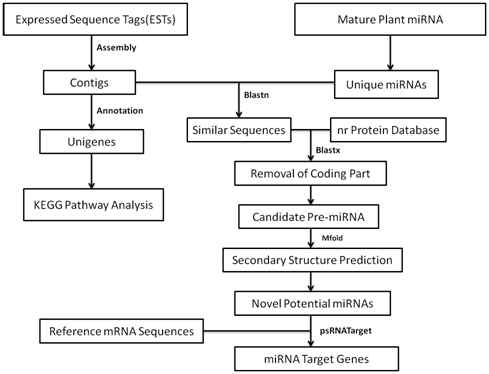
Figure 2 Diagrammatic representation of the scheme followed for the identification of miRNAs from turmeric ESTs.
Secondary structure prediction of miRNAs
The protein coding sequences were discarded while remaining non‒protein coding sequences were assessed for secondary structure using Mfold software.22 The other parameters were set as default. Potential candidates for pre‒miRNA were selected by subjecting them through following filter criteria given by Zhang et al.23
Prediction of potential turmeric miRNA target genes and ESTs
The target genes of miRNAs could be predicted through a homology algorithm24 on the basis of complementary binding of miRNAs (perfect or nearly perfect) to their target genes. For the prediction of the miRNA target psRNA target server was used with default parameters. The mRNAs of Curcuma longa were downloaded from NCBI. The potential miRNAs were served as query and were searched against mRNAs of Curcuma longa and also against assembled ESTs sequences. The parameters for prediction of the target included:
Phylogenetic analysis of mature miRNAs
Predicted miRNAs were searched against all known miRNAs using the standalone blast tool. The precursor miRNAs were collected from miRBase. The ClustalW was used to align collected precursor miRNA sequence along with the precursors of newly identified miRNAs. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 6.25 Evolutionary tree was constructed using maximum likelihood method.26
Annotation and pathway analysis of miRNA target genes
miRNAs were subjected to BLASTx search against non‒redundant protein sequence database of NCBI. GO terms were assigned to target genes using BLAST2GO 3.0 software (https://www.blast2go.com/blast2go‒pro/download‒b2g). Biological processes, cellular components and molecular functions associated with each GO term were examined. Furthermore, miRNAs were annotated using InterPro tool of Blast2GO software. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis27 was also performed using KASS server to find metabolic pathways and their networks (http://www.genome.jp/tools/kaas/).28
EST assembly and functional annotation of unigenes
A total of 12678 EST sequences were downloaded from dbEST of NCBI (http://www.ncbi.nlm.nih.gov/nucest/). The ESTs were further processed for assembly and functional annotation using CLC Genomics Workbench 8.0 (http://www.clcbio.com/products/clc‒genomics‒workbench/), and BLAST2GO21 suite, respectively. The EST assembly resulted into 2710 contigs, termed as unigenes after removing redundancy. The average read length of turmeric unigenes was 793 bp. The summary data of the EST assembly, clustering and annotation are presented in Table 1.
Item |
Number |
Total number of ESTs |
12678 |
Number of contigs |
2710 |
Total number of bases in unigenes |
2151623 |
Maximum length in contigs (bp) |
2445 |
Minimum length in contigs (bp) |
185 |
Average length of contigs (bp) |
793 |
Number of unigenes annotated |
1756 |
Total number of unannoted unigenes |
954 |
Table 1 Summary of Expressed Sequence Tags (ESTs) assembly and annotation in turmeric
The unigenes were further analyzed using BLAST2GO suite wherein similarity search was performed using (BLASTx against various public databases, followed by mapping and assignment of Gene Ontology (GO) terms to the unigenes (Figure 3). In similarity search, out of total 2710 unigenes, 2474 (91.3%) showed significant hits while 236 (8.7%) did not show any significant similarity. The unannotated unigenes may be considered as unique to Curcuma longa. Moreover, the unigenes of Curcuma longa showed the maximum similarity with Musa acuminata, followed by Elaeis guineensis, and Phoenix dactylifera (Figure 4).
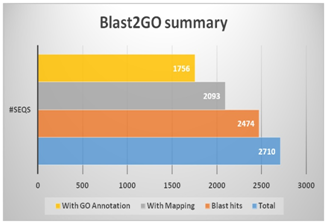
Figure 3 The summary of BLAST2GO results showing total number of sequences with number of blast hits, mapping and GO annotation.
Gene Ontology (GO) terms assigned to unigenes were found covering various molecular functions, biological processes, and cellular components (Figure 5). The further characterization showed that ion binding, heterocyclic compound binding, organic cyclic compound binding genes are most highly represented among the various molecular functions. The genes involved in transferase activity, small molecular binding and hydrolase activity also showed significant representation under molecular functions. In case of biological processes the genes involved in cellular process are markedly represented followed by metabolic and single‒organism processes. Further, cell part, membrane bound organelle and organelle part showed highest representation in the case of cellular components.
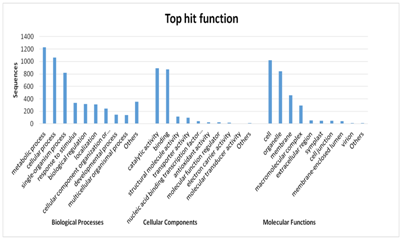
Figure 5 Gene Ontology (GO) terms assigned to turmeric unigenes among various categories representing molecular functions, biological processes, and cellular components.
The annotated unigene sequences of turmeric were mapped with reference pathways of Kyoto Encyclopedia of Genes and Genomes (KEGG). In total, 109 KEGG pathways were identified with biosynthesis of antibiotics being the most abundant followed by glycolysis/gluconeogenesis and purine metabolism pathways. This observation supported the traditional conception about turmeric of having medicinal values. In the classes of enzymes, the EC: 2‒transferase was maximum represented followed by EC: 1‒oxidoreductases, EC: 3‒hydrolases, EC: 4‒lyases, EC: 6‒ligases and, EC: 5‒isomerases.
Identification of miRNAs in turmeric ESTs
A total of 2710 unigenes were further processed for the identification of miRNAs. In case of plants, most of the mature miRNAs are found to be evolutionary conserved.2,5,29,30 In silico identification of miRNAs in ESTs has become one of the most conventional tools in recent times because of their evolutionary conserved behavior. In the present study, the computational and comparative genomics approach was used to identify conserved and novel miRNAs in assembled turmeric ESTs. In total, 8439 mature miRNAs of Viridiplantae (miRBase Release 21) were downloaded from miRBase (http://www.mirbase.org/cgi‒bin/browse.pl). After removing redundancy, 4647 unique miRNA were taken as reference and searched against 2710 unigenes, using locally installed BLAST tool version blast 2.2.30+(ftp://ftp.ncbi.nlm.nih.gov/blast). The parameters were set for BLASTn similarity search as: E value‒10, word size‒7 and window size‒7. The search yielded 102 potential mature miRNAs showing homology with 51 contigs/unigenes. The potential miRNA candidates had a length range of 16 nt to 22 nt, and the results are in agreement with previous studies in other plant species.5,31‒33 Further, BLASTx was performed to remove protein coding sequences out of potential miRNA candidates. None of the miRNA candidates was found to be present in the protein coding region, and thus all were retained for further analysis.
Mfold software was used to assign the secondary structures such as hairpin stem loops to selected candidate miRNAs. The various filters as mentioned in materials and methods section were employed to select the potential miRNAs. Out of 102, only four miRNAs fulfilled the criteria as a potential miRNA and are listed in Table 2. The Minimum Folding Free Energy (MFE) and Minimal Folding Free Energy Index (MFEI) are the two important criteria to determine the stability of hairpin loop secondary structure of precursor miRNA. According to Prabu et al.34 the value of MFE is inversely proportional to the stability of hairpin stem loop secondary structure of precursor miRNA, such that lower is the value of MFG, higher is the stability of secondary structure of precursor miRNA. The two values were also calculated in the case of turmeric precursor miRNA (Table 2). The values obtained suggested higher stability of candidate precursor miRNA identified in turmeric ESTs. The hairpin stem loop secondary structures of all the four identified mature and precursor miRNA of turmeric are represented in Figure 6A‒6D.
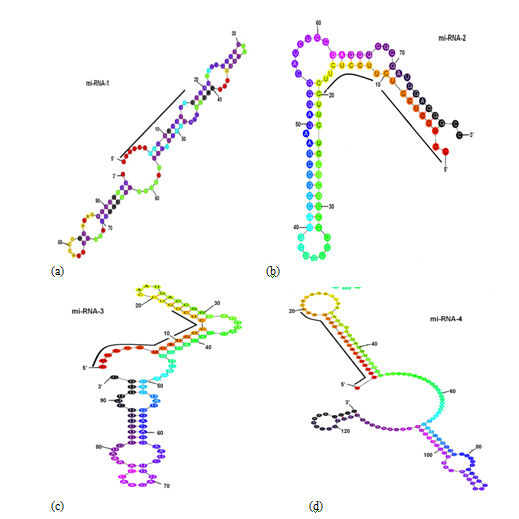
Figure 6 Secondary structure of pre-miRNAs
a) pre-miRNA-1
b) pre-miRNA-2
c) pre-miRNA-3, and
d) pre-miRNA-4
Source |
Homologous miRNA |
(G+C)% |
MFE(-∆G) |
MFEI |
Mature miRNA Sequence |
LM |
NM |
miRNA-1 |
slymiR5303/121 |
35.3 |
25.8 |
0.31 |
UUUUUGAAGAGUUCGAG |
17 |
0 |
miRNA-2 |
mtrmiR2673a/122 |
60 |
23.7 |
0.29 |
CCUCCUCCUCUUCCUCUUCC |
20 |
1 |
miRNA-3 |
mtrmiR5224a/122 |
64.71 |
27 |
0.33 |
CGUCCCUCAUGUCCUCG |
17 |
0 |
miRNA-4 |
mtrmiR5298d/124 |
30 |
27.9 |
0.33 |
AGAUGGAUAUGAAGAUGAAA |
20 |
0 |
Table 2 The novel miRNA identified in turmeric ESTs.
The various characteristics of novel identified miRNA are defined as; MFE, Minimum Folding Free Enrgy; MFEI, Minimum Folding Free Enrgy Index; LM, Length of Mature miRNA (nt); NM, Number of Mismatch between predicted and homologous miRNA
Zhang et al.5 suggested that in case of plants, the probability of finding a miRNA is one per 10000 ESTs. However, in our study, the presence of four miRNA in 12678 ESTs of turmeric, proved to be on the higher side. Other important features such as nucleotide percentage (A%,C%,G%,U%,A/U,G/C,A+U%) of newly identified miRNA in turmeric ESTs are summarized in Table 3. The average percentage of A+U remained 52.4% in miRNA precursor in turmeric, supporting the earlier findings of Zhang et al.5 In case of mature miRNA of turmeric, the U% dominated followed by G%,C% and A%, among the four nucleotides.
miRNA |
Mature miRNA Sequence |
miRNA Family |
A% |
C% |
G% |
U% |
A/U |
G/C |
A+U% |
miRNA-1 |
UUUUUGAAGAGUUCGAG |
slymiR5303/121 |
23 |
5.88 |
29.41 |
41.18 |
0.57 |
5 |
64.7 |
miRNA-2 |
CCUCCUCCUCUUCCUCUUCC |
mtrmiR2673a/122 |
0 |
60 |
0 |
40 |
0 |
0 |
40 |
miRNA-3 |
CGUCCCUCAUGUCCUCG |
mtrmiR5224a/122 |
5.8 |
47.07 |
17.65 |
29.41 |
0.2 |
0.375 |
35.29 |
miRNA-4 |
AGAUGGAUAUGAAGAUGAAA |
mtrmiR5298d/124 |
50 |
0 |
30 |
20 |
2.5 |
0 |
70 |
Table 3 Some important features of newly identified miRNA in turmeric
miRNA target prediction
In plants, miRNAs regulate gene expression via translational repression, mRNA cleavage and deadenylation, and target functional genes involved in various physiological processes including growth, development, stress stimulus, defense, etc.35 MiRNAs identify their target mRNAs through perfect or near‒perfect complementarity and initiates cleavage.2,36 A homology based tool, psRNA Target server was used for the prediction of turmeric miRNA target mRNAs. To identify the potential mRNA targets in RefSeq and assembled ESTs of turmeric, the four newly identified miRNA were used as query. A total of 10 targets for these newly identified four mature miRNAs of turmeric were found. Out of 10 targets, six were of miRNA‒2 and rest four represented miRNA‒4 (Table 4).
miRNAs |
Target accession no. |
Target Protein |
Inhibition |
Target Activity |
GO Annotation (Biological Process) |
miRNA-2 |
gi|396118857 |
beta-glucosidase 18-like |
Cleavage |
Starch and sucrose metabolism |
Carbohydrate metabolic process |
miRNA-4 |
gi|396132539 |
dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 |
Translation |
Cystein and methionine metabolism |
L-methionine biosynthetic process from methylthioadenosine, regulation of cell division, oxidation-reduction process |
miRNA-4 |
gi|396155903 |
protein chloroplastic |
Translation |
Phenylalanine metabolism |
Response to oxidative stress, chloroplast organization, protein import into chloroplast stroma, oxidation-reduction process |
miRNA-4 |
gi|396150737 |
14 kda zinc binding protein |
Translation |
Caretenoid biosynthesis |
Sulpher compound metabolic pathway, purine ribonucleotide metabolic pathway |
miRNA-4 |
gi|396155903 |
protein chloroplastic |
Translation |
Phenylalanine metabolism |
Response to oxidative stress, chloroplast organization, protein import into chloroplast stroma, oxidation-reduction process |
miRNA-2 |
gi|396123884 |
glucose-6-phosphate-1-epimerase |
Cleavage |
Glycosis/gluconeogenesis pathway |
Carbohydrate metabolic process |
miRNA-2 |
gi|396118857 |
beta-glucosidase 18-like |
Cleavage |
Starch and sucrose metabolism |
Carbohydrate metabolic process |
miRNA-2 |
gi|396123699 |
gdsl esterase lipase at 4g01130 |
Cleavage |
Starch and sucrose metabolism |
Oxidation-reduction process, 9,9'-di-cis-zeta-carotene desaturation to 7,9,7',9'-tetra-cis lycopene |
miRNA-2 |
gi|396118857 |
beta-glucosidase 18-like |
Cleavage |
Cyanoamino acid metabolism |
Carbohydrate metabolic process |
miRNA-2 |
gi|396123884 |
glucose-6-phosphate-1-epimerase |
Cleavage |
Glycosis/gluconeogenesis pathway |
Carbohydrate metabolic process |
Table 4 List of potential target genes of two newly identified turmeric miRNA (miRNA-2 and miRNA-4)
When turmeric contigs/unigenes were subjected to analysis of target of these miRNAs, 38 targets were identified including 33 contigs/unigenes as target of miRNA‒2 while miRNA‒4 targeted 5 contigs/unigenes. No target was found for miRNA‒1 and miRNA‒3, suggesting these miRNAs may have some novel targets hitherto unreported. The major biological processes performed by the target gene of miRNA‒2 are carbohydrate metabolism and oxidation‒reduction processes. On the other side, the target genes of miRNA‒4 are involved in diverse processes such as L‒methionine biosynthetic process from methylthioadenosine, regulation of cell division, oxidation‒reduction process, response to oxidative stress, chloroplast organization, protein import into chloroplast stroma, oxidation‒reduction process, sulpher compound metabolic pathway, and purine ribonucleotide metabolic pathway.
Phylogenetic analysis of identified miRNAs
The identified precursor miRNAs were subjected to multiple sequence alignment using Clustal W. This analysis helps to study relationship of precursor miRNAs with that of other members of the same family. Their evolutionary relationships were established using maximum‒likelihood method in MEGA. Conserved nature of pre‒miRNA and mature miRNA in distantly related species has already been reported by various researchers including Zhang et al.5 miRNA‒1 is closely related to slymiR5303 while miRNA‒2, miRNA‒3 and miRNA‒4 showed maximum similarity with mtrmiR2673a, mtrmiR5224a and mtrmiR5298d, respectively. Among these four, miRNA‒2 and miRNA‒3 showed phylogenetically more closeness compared to miRNA‒1 and miRNA‒4. Their bootstrap values can be seen from the phylogenetic tree (Figure 7).
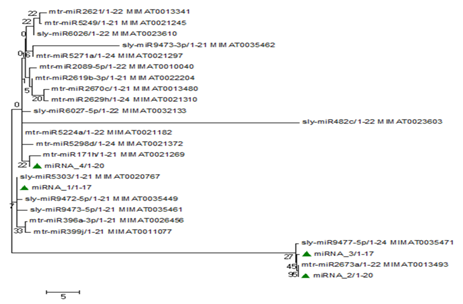
Figure 7 Phylogenetic tree prepared using maximum likelihood method showing relationship between newly identified pre-miRNAs of turmeric with their closely related families.
GO term annotation and KEGG pathway analysis
To understand the function of the newly identified miRNAs, the targeted genes were subjected to assign Gene Ontology (GO) terms. The targeted genes were assigned to regulatory network in all the three terms i.e. molecular function, biological process and cellular component (Figure 8). In molecular function, genes involved in oxidoreductase activity are most abundant followed by hydrolase activity, whereas in biological process, genes for primary metabolic process and single organism metabolic process are highly represented. Cell part and organelle part genes are most represented in the cellular component category.
The turmeric (Curcuma longa L.) is known for its medicinal and culinary value for long. In this study, four novel miRNAs and their targeted genes in turmeric have been identified that play significant role in metabolic processes. The identification of 35 target genes suggested that these miRNAs regulate multiple gene expression by modulating many regulatory molecules like transcription factors, enzymes and secondary messengers. The study will provide a lead in the direction of understanding the miRNA regulatory patterns in turmeric as well as in other important non‒model plants.
None.
The author declares that there is none of the conflicts.

©2018 Rai, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.