Journal of
eISSN: 2572-8466


Research Article Volume 2 Issue 4
1DBT-ICT Centre for Energy Bioscience, India
2Aberystwyth University, United Kingdom
Correspondence: Annamma A Odaneth, DBT-ICT Centre for Energy Biosciences, Institute of Chemical Technology, Matunga, Mumbai-400019, India
Received: October 27, 2016 | Published: March 9, 2017
Citation: Rodrigues VJ, Onime L, Huws SA, et al. Diversity of ulvan and cellulose depolymerizing bacteria associated with the green macroalgae Ulva Spp . J Appl Biotechnol Bioeng. 2017;2(4):136-142. DOI: 10.15406/jabb.2017.02.00037
Ulva spp. is a green macroalgae with high cell wall polysaccharide content. Ulvan and cellulose are the major polysaccharides present in Ulva spp. breakdown products of which have potential applications in the biofuel and fine chemical industry. Bacteria attached to Ulva spp. potentially possess enzymes that have the capability to depolymerize these polysaccharides. Ulvan and cellulose depolymerizing bacterial communities associated with fresh and rotting green macroalgae Ulva spp. collected from Aberystwyth shore (Wales, United Kingdom) were studied with the aim of understanding the breakdown of these polysaccharides in nature. Polysaccharide breakdown capability was investigated by the screening of these organisms for various enzymes involved in the breakdown of cellulose and ulvan. Positive organisms were identified using the 16S rRNA homology approach by amplification and sequencing of their 16S rRNA gene using universal primers, followed by homology analysis using NCBI BLAST. Phylum Proteobacteria with class Gammaproteobacteria predominated the community and was represented by species of orders Oceanospirillales, Vibrionales, Alteromonadales and phylum Bacteroidetes was represented singly by a species belonging to order Flavobacteriales. A synergistic mechanism was observed towards the breakdown of these polysaccharides with different sets of organisms contributing towards different enzymes required for the complete saccharification of ulvan and cellulose.
Keywords: ulva spp, ulvan, cellulose, 16S rRNA, ulvan lyase, carbohydrate sulfatase, endocellulase, exocellulase, β-glucosidase
rRNA, ribosomal ribonucleic acid; DNA, deoxyribonucleic acid; CMC, carboxymethyl cellulose; UV, ultraviolet; PCR, polymerase chain reaction
Marine microbial ecology has received considerable attention in the past decade owing to the fact that microbes regulate the functioning of the marine ecosystem.1 Host- associated bacterial communities play an important part in the life cycle of marine flora, yet our understanding of these microbial communities is limited with respect to their diversity and the role they play in the life cycle of the host they associate themselves to. These microbes are assumed to hold the key to utilize the various resources that the marine habitat has to offer. Enzymes such as proteases, lipases, chitinase, chitosanases, alginate lyases, agarases, carrageenases, cellulose and hemicellulose hydrolases, amylases, fucodian lyases and more, of marine bacterial origin can be utilized for breakdown of the various polymers found in the marine environment as well as polymers of terrestrial origin. These enzymes are also known to be tolerant to extreme salinity, temperature and pressure owing to the conditions they are exposed to in the environment their hosts thrive in.2 Macroalgae are an important part of the marine ecosystem and hold tremendous potential for commercial exploitation to produce high value chemical and biofuels. Worldwide researchers are focusing on utilization of biomass of varied macroalgae for biofuel, food, biomedical and in the fine chemical industry. The green macroalgae Ulva spp. are interesting seaweed whose cell wall polysaccharides are being investigated as a sustainable source of third generation biofuels and other value added compounds.3 Ulva spp. is known for their quick proliferation and high polysaccharide content i.e around 65% of its dry weight which mainly comprises of ulvan and cellulose.4 They are also known to cause green tides around the world.5,6 Despite their enormous potential they are currently not commercially utilized and research towards utilization of this biomass needs to be carried out.
The first step towards exploiting the commercial potential of polysaccharides in Ulva spp. involves their depolymerization. Cellulose has homologs in terrestrial biomass, and the depolymerization of cellulose has been greatly understood owing to years of research. However, enzymes available for the depolymerization of cellulose obtained from terrestrial biomass cannot be used for the cellulose present in Ulva spp. due to the high salinity of the biomass. Ulvan on the other hand has no known homologs in terrestrial plants and the breakdown of ulvan has not yet been completely understood. The key to understanding the breakdown of these polysaccharides is to study their breakdown in nature. Macroalgae are known to harbor diverse organisms associated on its surface which participate in various stages of the lifecycle of the host.1 Considering that the process of surface colonization and biofilm formation by bacteria are influenced by the type of carbohydrates available in the environment,7 the surface of Ulva spp. itself is the best possible source of organisms than can degrade the cell wall polysaccharides found in this macroalgae. Organisms know to colonize Ulva spp. according to previous studies belong to genus Pseudoalteromonas,8-11 Phaeobacter,8,11 Shewanella, Dokdonia, Acinetobacter, Microbacterium 12, Alteromonas, Vibrio ,10,11,13 Cobetia .14 Sulfitobacter, Halomonas, Roseobacter,15 Marinomonas, Bacillus 16 are also known to be associated with Ulva spp. and play a role in their morphology development. This study focuses on screening of organisms associated with fresh and rotting green macroalgae Ulva spp. for enzymes that are capable of breaking down the two major polysaccharides ulvan and cellulose. This will help us understand the microbial diversity as well as the synergism between these diverse organisms which exists in the natural environment towards the breakdown of these polysaccharides.
Sample collection
Fresh and rotting Ulva spp. samples were collected from two different sites on Aberystwyth shore (52°24′ N; 4°04′ W), Wales, United Kingdom. Samples were collected by hand during low tide from the rocky areas at the two extremes of the northern part of the shore and are referred to as Site 1 and Site 2 hereafter. Collected samples were sealed in a zip lock bag and transported immediately to the lab. The algal surface was rinsed once with sterile fresh water and used for isolation of organisms.
Isolation of bacterial strains
Isolation was carried out using two different methods. Algal thallus from the different samples were cut and placed directly on plates containing DifcoTM Marine Agar 2216. In the second method a piece of algal thallus was submerged in 5 ml of autoclaved sea water and vigorously vortexed, 100µl of this was spread on DifcoTM Marine Agar 2216 plates. The inoculated plates were maintained at a temperature of 15°C until colonies were observed. Colonies were picked up using a sterile loop and sub cultured until single colonies were obtained. Single colonies were grown in tubes containing DifcoTM Marine Broth 2216 17 and labelled sequentially.
Screening for enzyme activity
Isolated organisms grown in broth were aliquoted in 384 well plates, replicated on agar based assay plates and screened for five enzymes i.e ulvan lyase and carbohydrate sulfatase required for the saccharification of ulvan and exocellulase, endocellulase and β-glucosidase required for the saccharification of cellulose.
Ulvan lyase screening: Screening for organisms with ulvan lyases was carried out as described by Collén PN et al. 17 Briefly 1g/L of ulvan (Elicityl oligotech, France) was added to a mixture of 100 mM Tris- HCl pH 8.5 (Sigma-Aldrich, United Kingdom) and 200 mM NaCl (Sigma-Aldrich, United Kingdom) and DifcoTM Marine Broth 2216. This medium was solidified with 1% agarose (Bio-Rad, United Kingdom). Organisms were deposited on the plate using a 384 well plate replicator and incubated at 15°C for two 48 hours. The plate was developed using 0.05% Ruthenium Red (Sigma-Aldrich, United Kingdom) for 10-15 minutes. Activity was detected as clear spots against a pink background.
Carbohydrate Sulfatase screening: Carbohydrate sulfatase possessing bacteria were screened for by a method described by Byun DS et al. 18. DifcoTM Marine Agar 2216 plates were prepared containing 5 mM p-Nitrophenyl Sulfate (Sigma-Aldrich, United Kingdom). Organisms were deposited on the plate using a 384 well plate replicator and incubated for 48 hours at 15OC. Activity was detected by the appearance of a yellow halo around the positive organisms.
Cellulases screening: Screening for cellulases was done using techniques described by Hurek BR et al. 19, Organisms were grown on DifcoTM Marine Agar 2216 plates in the 384 well plate format for 48 hours. An overlay consisting of 20 ml of pH 7.0 0.05 M potassium phosphate (Sigma-Aldrich, United Kingdom) buffered agarose (0.7%) (Bio-Rad, United Kingdom) containing 0.5mg/ml of 4-Methylumbelliferyl-3-cellobioside (Sigma-Aldrich, United Kingdom) for exocellulases, 1% CMC sodium salt of medium viscosity (Sigma-Aldrich, United Kingdom) for endocellulases and 5 mM 4-Nitrophenyl-β-D-glucopyranoside (Sigma-Aldrich, United Kingdom) for β-glucosidase screening was poured on the plates. These plates were incubated with the overlays at 15 OC for approximately 6 hours. Plates to be screened for exocellulases were exposed to UV light. Active colonies were identified by the presence of fluorescence under UV. Plates to be screened for endocellulases were developed with 1% w/v Congo red (Sigma-Aldrich, United Kingdom and activity was identified by appearance of a clear yellowish halo on a red background. β-glucosidase activity was detected by appearance of yellow halo around the positive clones.
Identification of positive organism
Organisms were identified on the basis of their 16S rRNA gene sequence similarity. Liquid cultures (50µl) of organisms positive for tested activities were boiled at 100°C for 10 minutes in PCR plates. Boiled cultures were then centrifuged at 10,000X g for 10 minutes and the supernatant was used for PCR amplification of the 16S rRNA gene. The PCR reaction was performed in a 25 µl volume and contained 1 µM of each primer, 1× MyTaq Red Buffer (Bioline, United Kingdom), 0.5unit of My Taq red DNA polymerase (Bioline, United Kingdom), 16.4 µl of ultra-distilled H2O and 50-100 ng of template DNA. Universal primers 27F (5’AGAGTTTGATCMTGGCTCAG-3’) and 1389R (5’ACGGGCGGTGTGTACAAG-3’) were used for amplification. The amplification reactions were carried out in an Applied Biosystems 2720 Thermal Cycler using the following program: 4 min at 94°C, 45 sec at 94°C, 45 sec at 55°C, 1.30 min at 72°C for 30 cycles, with an additional elongation step at 72°C for 7 min.20 PCR products were purified using a QIAGEN QIAquick PCR purification kit. Sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI3130xl DNA sequencer at the Sequencing facility at Aberystwyth University. Organisms were identified on the basis of homology of the 16S rRNA gene to gene sequences of known organisms using the NCBI-BLAST tool.
Isolation of bacterial strains
A total of 849 organisms were isolated from fresh and rotting green macroalgae Ulva spp. of which 367 were isolated from the fresh algal thallus and 482 were isolated from the rotting algal thallus. The organisms isolated from algal thallus sampled at different sites and using different methods can be categorized as depicted in Table 1.
Screening of strains for polysaccharide breakdown enzyme activity
Of the total 849 strains screened for five enzymes required for the saccharification of cellulose and ulvan, 235 tested positive for one or more enzymes. Ulvan lyases were present in 166 strains, while carbohydrate sulfatases were present in 26 strains, β-glucosidases in 69 strains, endocellulases in 70 strains and exocellulases in 34 strains Figure 1. Illustrates the results of the screenings carried out for detecting the activity of the above mentioned enzymes. Venn diagram indicating the overall number of organism with single activity as well as number of organisms with more than one of the five enzyme activities screened for is illustrated in Figure 2.
Isolation method |
Number of Isolated |
||
Site 1 |
Site 2 |
Total |
|
Rotting-Vortexed |
148 |
123 |
271 |
Rotting-Directly Plated |
123 |
88 |
211 |
Fresh-Vortexed |
82 |
131 |
213 |
Fresh-Directly Plated |
50 |
104 |
154 |
Total |
403 |
446 |
849 |
Table 1 Organisms isolated from different study sites against different isolation methods.

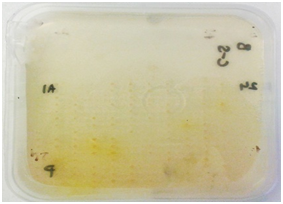
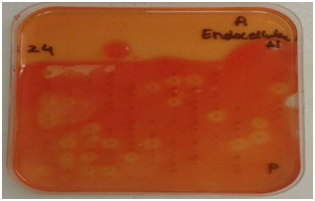

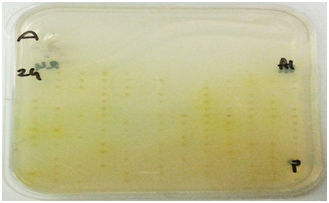
Figure 1 Plate screening assays for enzyme activity.
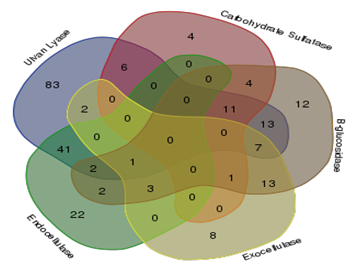
Figure 2 Venn diagram showing distribution of organisms having individual activities and more than one activity.
Identification of the isolates
Isolates were identified on the basis of their 16S rRNA gene sequence similarity. Phylum Proteobacteria with class Gammaproteobacteriapredominated the colonizing communityand its diversity was represented by genus Cobetia (9 isolates), Halomonas (20 isolates) and Zymobacter (1 isolate)of family Halomonadaceae, Order Oceanospirlalles; genus Vibrio (83 isolates), Photobacterium (1 isolate) and Alivibrio (1 isolate) of family Vibrionaceae, order Vibrionales; genus Moritella (1 isolate)of family Moritellaceae, order Alteromonadales; genus Pseudoalteromonas (14 isolates)and Psychormonas (1 isolate) of family Pseudoalteromonadaceae, order Alteromonadales and phylum Bacteroidetes was singlyrepresented bygenus Winogradskyella offamily Flavobacteriaceae order Flavobacteriales, while 103 isolates could not be identified. The isolated species belonging to respective genus and those from every genus positive for the various activities is summarised in Table 2.
Genus |
Number of isolates |
Number of Organisms Positive for Various Activities |
||||
Ulvan Lyase |
Carbohydrate Sulfatase |
Exocellulase |
Endocellulase |
β- glucosidase |
||
Vibrio |
83 |
49 |
24 |
6 |
0 |
52 |
Halomonas |
20 |
19 |
0 |
0 |
0 |
1 |
Cobetia |
9 |
9 |
0 |
0 |
0 |
0 |
Psychromonas |
1 |
1 |
0 |
0 |
0 |
0 |
Pseudoalteromonas |
14 |
10 |
0 |
0 |
5 |
1 |
Zymobacter |
1 |
1 |
0 |
0 |
0 |
0 |
Moritella |
1 |
1 |
0 |
0 |
0 |
0 |
Photobacterium |
1 |
1 |
0 |
0 |
0 |
0 |
Winogradskyella |
1 |
1 |
0 |
0 |
0 |
0 |
Aliivibrio |
1 |
1 |
0 |
0 |
0 |
0 |
Unidentified |
103 |
73 |
3 |
10 |
55 |
14 |
Total |
235 |
166 |
27 |
16 |
60 |
68 |
Table 2 Number of organisms identified from each genus and the activities they possess.
Bacterial diversity associated with Ulva spp. demonstrating polysaccharide breakdown activity was systematically assessed in this study. Data indicated that the organisms predominantly belonged to phylum Proteobacteria and class Gammaproteobacteria whileonly a single organism belonging to phylum Bacteroidetes and class Flavobacteria was identified. Zhang X et al.21 have observed that Gammaproteobacteria are the dominant class in environments where algal blooms occur and are represented mostly by Vibrios. Results of the current study are in agreement to this observation, with 35% of the positive organisms and 80% of identified organisms belonging to genus Vibrio. Results of this study are also supported by Edwards JL et al.22 who investigated bacterial colonies on marine biofilms having cellulose degrading activity, and observed that class Gammaproteobacteria predominated the colonies. While they also observed organisms belonging to phylum Bacteroidetes in abundance, our results indicate only a single organism from this phylum. Recent research has indicated that bacteria belonging to phylum Bacteroidetes are specialists in degradation of macromolecules with a large number of glycoside hydrolases and proteases.23,24,25 The under representation of Bacteroidetes in this study could possibly be due to the use of a culture based approach, which led to selective isolation of Proteobacteria or due to a bias in the universal primers used for amplification of the 16S rRNA genes, considering that 103 organisms could not be identified.
Vibrios constituted a major portion of the organisms isolated from both the fresh and the rotting thallus. Studies have indicated that Vibrio spp. play a pivotal role in degradation of polysaccharides found in the marine environment. Enzymes for polysaccharide breakdown isolated from Vibrio spp. include a novel endo type β- agarase involved in the degradation of agarose,26,27,28 alginate lyases,29,30,27 chitinases,31 β-1,3-Xylanase,32 laminarinase,33 endo-β-1,4-glucanase,34 carbohydrate binding modules linked to a β-1,3-Xylanase ,35fucoidanase and fucoidan sulfatase,36 k-carrageenase 37 and cellulase 38-40. This study reports for the first time the detection of ulvan lyase and carbohydrate sulfatase activity in organisms belonging to Vibrio. Exocellulase and β-glucosidase activity was also detected in the isolated Vibrio species. This indicates that Vibrios have diverse sets of enzymes that are capable of depolymerizing a variety of carbohydrates in the marine environment. It can be hypothesized that the expression of genes that code for these enzymes is regulated by the availability of carbohydrates in the environment. Vibrios have also been isolated from the gut of marine herbivores such as Haliotis discus,41,42 Sparus aurata,43 Aplysia kurodai 44that feed on seaweed. The source of large number of Vibrios observed in this study could possibly be the feces of such marine herbivores present in the environment. Genomes of some of these Vibrio species have been sequenced as part of our further study and will help us understand the depolymerization of polysaccharides in the marine environment.
Halomonas is the next most dominant population of bacteria identified in our study, representing 8% of positive organisms and 19% of the identified organisms. Several studies have reported polysaccharide degrading enzymes identified and isolated from bacteria belonging to the genus Halomonas. Enzymes identified in previous studies include, α-D-glucosidase, β-D-glucosidase and β-D-galactosidase,45 carboxymethylcellulase (CMCase), xylanase, lipase, amylase and pullulanase. Enzymes derived from Halomonas are also known to work under extreme pH and salinity ranging from pH 5.0-11.0 and NaCl concentration of 0-15%,46 this makes enzymes derived from bacteria belonging to this genus ideal for depolymerization of polysaccharides from marine biomass. Halomonas strains isolated in this study mostly displayed ulvan lyase activity with only a single strain displaying β-glucosidase activity. None of the isolated strains display carbohydrate sulfatase activity. We can thus hypothesize that organisms belonging to genus Halomonas either metabolize the sulfated breakdown products of ulvan or depend on other organisms in the environment to release enzymes for the removal of sulfates present in ulvan. Pseudoalteromonas is the third most dominant population of bacteria found on the surface of Ulva spp. Carbohydrate breakdown activity has been reported in the past in organisms belonging to genus Pseudoalteromonas. Enzymes identified and isolated in other studies include carrageenan sulfatase,47 β-(1-4) agarase,48,49 λ- Carrageenase,50 k- Carrageenase,51 Cellulases.52 Ulvan lyase activity has also been reported in Pseudoalteromonas spp..53 This study confirms the ulvanolytic activity of bacteria belonging to genus Pseudoalteromonas. Further study on the ulvan lyases produced by the organism isolated in this study will help us determine if the enzyme is the same or diversity exists in the enzyme produced by different species from the same genus.
Nine isolates belonging to genus Cobetia and single isolates belonging to genus Alivibrio, Winogradskyella, Psychromonas, Moritella, Photobacterium, and Zymobacter were isolated and identified in this study. All of these possess only ulvan lyase activity. Data on the other polysaccharide degrading activity possessed by the above organisms is scarce. It is possible that these organisms are poorly represented on the macroalgal thallus because of the dominance of Vibrios, Pseudalteromonas and Halomonas. Considering the different polysaccharide breakdown activites that have been reported in literature for the various genus of bacteria isolated from the thallus of Ulva spp. in our study we can also conclude that organisms belonging to genus Vibrio, Pseudoalteromonas and Halomonas, have the ability to degrade almost all of the different polysaccharides present in the macroalgae found in the marine environment. The inability to amplify the 16S rRNA of 103 of the positive organisms could be attributed to two possibilities a bias in the universal primers 27F and 1389R towards amplification of only Proteobacteria over other phylum or the isolated organisms are possibly archaea or eukaryotes and not bacteria. Further work needs to be carried out to determine the identity of these organisms. Universal primers for the amplification of conserved genes of archaea, eukaryotes and other phylum belonging to bacteria can be used to try and identify these organisms.
Result of this study help us to conclude that that a synergistic mechanism exists in nature with different organisms contributing to the array of enzymes required for the depolymerization of the carbohydrates present in Ulva spp. Kopel M et al.53 in their study on polysaccharide degrading bacteria have observed the presence of polysaccharide utilization loci involved in degradation of sulfated polysaccharides indicating the possibility of such regions being present in the genomes of the organisms isolated in this study. In depth analysis of the genomes of isolated organisms will further help us understand their contribution towards breakdown of polysaccharides present in the environment. Application of this knowledge towards designing of enzyme systems for saccharification of Ulva spp. biomass will involve development of a concoction of enzymes sourced from several organisms or genetic modification of a single organism for production of all of the required enzymes.
This study was funded by Department of Biotechnology, Ministry of Science and Technology, Government of India and Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
The author declares no conflict of interest.

©2017 Rodrigues, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.