Journal of
eISSN: 2572-8466


Research Article Volume 2 Issue 1
1Department of Biochemistry and Microbiology, Sao Paulo State University, Brazil
2Graduate Program of Biotechnology and Environmental Monitoring PPGBMA, Federal University of Sao Carlos (UFSCar), Brazil
3Cell and Molecular Biology Laboratory, University of Sao Paulo, Brazil
Correspondence: Acacio Aparecido Navarrete, Federal University of Sao Carlos Rod, Jo?o Leme dos Santos km 110Sorocaba, SP, 18052-780, Brazil, Tel 55019-98141-6055
Received: October 28, 2016 | Published: January 26, 2017
Citation: Becker ACF, Neves FA, Angelis DDF, et al. Differential occurrence of heterotrophic bacteria to specific physicochemical characteristics of oil refinery wastewater and adjacent water bodies. J Appl Biotechnol Bioeng. 2017;2(1):18-23. DOI: 10.15406/jabb.2017.02.00020
This study used a multi-analytical approach based on traditional microbiological methods of cultivation and isolation of heterotrophic bacteria in laboratory, associated with partial sequencing of the gene 16 rRNA of 80 heterotrophic bacterial cultures isolated by plating in PCA media and physicochemical analysis of samples from oil refinery wastewater treatment and adjacent water bodies. For the isolation of the bacterial cultures and physicochemical determinations, wastewater samples were collected at the Paulinía’s Refinery (REPLAN/Petrobras), located in the state of São Paulo, Brazil, from input point of the raw effluent for biological treatment and output point of the effluent from stabilization pond; water samples were collected from adjacent water bodies at the draw point of water for wastewater treatment (Jaguari River) and upstream the disposal of the effluent (Atibaia River). Chemical determinations included dissolved oxygen (DO) rates, chemical oxygen demand (COD), biochemical oxygen demand (BOD), chlorides, calcium carbonate acidity, alkalinity, ammonia, nitrite, nitrate, dissolved ions, sulfides, oil and grease and physical determinations covered pH measurements, conductivity, temperature, transparency, settleable solids, soluble and suspended solids, volatile matter, remaining fixing material, apparent color and turbidity. Based on network approach, theoretical relationships were shown involving Carnobacteriaceae and Aeromonadaceae and physicochemical characteristics of the wastewater and water samples. Carnobacteriaceae showed positive theoretical relationships with COD, BOD, nitrate, chloride, temperature, conductivity and apparent color and negative relationships with dissolved oxygen. Positive theoretical relationships were also shown between Aeromonadaceae and the DO rate and nitrate, while negative relationships were shown between the same bacterial family and the rates of COD, BOD, chlorides, ammonia, volatile matter, alkalinity, apparent color, acidity and conductivity. The results raise the possibility for the investigation of Carnobacteriaceae and Aeromonadaceae families as potential indicators for the evaluation of the efficiency of methods adopted for the biological treatment and disposal of residues derived from oil refining.
Keywords: microbial indicators, network approach, carnobacteriaceae, aeromonadaceae
DO, dissolved oxygen; COD, chemical oxygen demand; BOD, biochemical oxygen demand
Petroleum refining is a process which consumes an average of 246 to 340 L of water per crude oil barrel and generates effluent that turns out to be 0.4 to 1.6 times the amount of processed oil.1,2 Environmental pollutants are constituent parts of this process, such as cyanide, heavy metals, phenols, sulfides, ammonia and polycyclic aromatic compounds. The residuary water produced in oil refining plants is a threat to quality of water bodies downstream of the discharge point. The presence of organic compounds and other contaminants tend to rise levels of biochemical oxygen demand (BOD) and chemical oxygen demand (COD), reducing the amounts of available oxygen for organisms living in that environment.2-4 Microorganisms may be single-celled or multi-cellular, with prokaryotic organisms consisting of a cell nucleus and the other membrane bound organelles. This structure ensures high sensibility to environmental variation for the prokaryotes whose distribution, abundance and variety of different species or groups can respond differently to human-induced changes.5,6 This characteristic of the prokaryotes opens the possibilities to investigate them as potential indicators for environmental changes. The counting of heterotrophic bacteria is widely used as an indicator of water quality, being the microorganisms detected by propagation in nonselective media.7 However, despite the practicality of this culture-dependent approach, an accurate taxonomic identification of the bacterial community grown on media culture is made possible by extracting and analyzing its total nucleic acids.
Analysis of water physical and chemical properties such as pH, conductivity, temperature, turbidity, dissolved oxygen, biochemical oxygen demand, chemical oxygen demand and others is commonly used by laboratories to determine water quality. Despite the importance of diagnosis based on these water properties, the microorganisms should also integrate this analysis. However, new studies focusing on water microbial communities are necessary to identify microbial indicators that hold potential for use as bio indicators of disturbance in natural water bodies after effluent disposal. Because of the substantial effects that residuary water produced in oil refining plants may have on the chemical and physical characteristics of natural water bodies and the possibility of prokaryotes indicate the water quality status, we would like to obtain better insight into the ecological characteristics of heterotrophic bacterial community recovered from two rivers (Jaguari and Atibaia) involved in the treatment and discharge, respectively, of the effluent produced at the Paulinia Refinery (REPLAN-Petrobras), state of São Paulo, Brazil.8 For this purpose, we performed the 16S rRNA gene sequencing based on bacterial colonies grown in culture medium after inoculation of water samples. In addition, water physical and chemical properties were analyzed in all sampling points and a network approach was used to integrate bacterial community composition data with physicochemical properties in order to identify bacterial groups that hold potential for use as indicators for quality and effectiveness of the biological treatment and effluent discharge methods used in oil refineries.
Sampling sites and water and effluent sampling
This study was performed at Paulínia Refinery (REPLAN/Petrobras) and two adjacent rivers. Wastewater samples were collected at the refinery from input point of the raw effluent for biological treatment, site 1 and output point of the effluent from stabilization pond, site 2. Water samples were collected from Jaguari River at the draw point of water for wastewater treatment (S 22°41’48’’/W 47°08’59’’), site 3 and from Atibaia River, upstream the disposal of the effluent (S 22°41’48’’/W 47º07’22’’), site 4. Sampling sites are shown in Figure 1. The samples collection and preservation were according to CETESB.9 Atibaia River belongs to class 2 according to CONAMA’s 357/05 Resolution. In this class, water can be used for human consumption after simplified treatment and recreational activities such as swimming and agricultural irrigation for vegetables and fruits growing close to the soil.
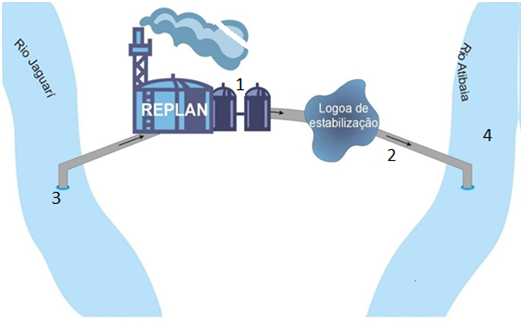
Figure 1 Scheme of the sampling sites location at Paulínia Refinery (REPLAN/Petrobras) and adjacent rivers.
Site 1: input of the biological treatment (raw effluent).
Site 2: output of the stabilization pond.
Site 3: drawn point at Jaguari River of water for wastewater treatment.
Site 4: upstream the discharge of the effluent in the Atibaia River.
Analysis of wastewater and water physicochemical factors
Physicochemical factors were analyzed based on samples collected from the four sampling sites according to Eaton et al.10 Physical factors consisted of pH, conductivity, temperature, transparency, settleable solids, suspended and soluble solids, volatile matter, remaining fixing material, apparent color and turbidity. Chemical factors encompassed dissolved oxygen (DO), chemical oxygen demand (COD), biochemical oxygen demand (BOD), chloride, CaCO₃ acidity, alkalinity, ammonia, nitrite, nitrate, sulfide, cyanide, phenols, oils and greases.
Inoculation of wastewater and water, and growth and isolation of heterotrophic bacterial colonies
An accurate aliquot (1 mL) from each of the two wastewater and two water samples was suspended in 9 mL sterile water and mixed using a Teflon-coated magnetic bar for 4 min at approximately 200 rpm. The resulting suspensions were serially diluted in 10-fold steps by the addition of 1 mL of the previous dilution to 9 mL sterile water, stirring for 5 min between dilutions. Volumes of 0.1 mL from the 10-1 dilution of the sample were spread onto plates of Plate Count Agar (PCA) medium using a sterile glass spreader. Individual colonies were picked from the plates and preserved in 10% glycerol at -80°C. Nutrient broth liquid medium (M002 Himedia™ Laboratories, Mumbai, India) was used for isolation of bacterial colonies. Therefore, 39.0 g of the culture medium were dissolved in 3 L of distilled water. Volumes of 30 mL of the solution were poured into 50 mL Falcon tubes. The tubes containing the medium were sterilized for 15 minutes in autoclave at 120°C and 1 atm pressure. A total of 80 bacterial colonies was inoculated into nutrient broth medium using sterile swabs in a bio-safety cabinet under aseptic conditions. The Falcon tubes containing the inoculums were maintained at 36°C for seven days (i.e. at a representative temperature for all the four sampling sites defined based on the real range of temperature). A volume of 0.5 mL of Brain Heart Infusion (BHI) broth was added into the tubes that had not shown any growth until five days after inoculation. The growth was suspended 7 days after inoculation and DNA was isolated from each of 80 bacterial cell suspensions.
DNA isolation from bacterial cell suspensions
Falcon tubes containing bacterial cell suspensions were centrifuged at 10.000 rpm for 1.5 min to concentrate the cells. After that, the supernatant was discarded and a volume of 1.8 mL of bacterial cells was added to MicroBead Tubes from the UltraClean Microbial DNA Isolation™ kit (MoBio Laboratories, Carlsbad, CA, USA). The cells were lysed by horizontal agitation at 10,000 x g for 30 sec and DNA was isolated according to manufacturer’s instructions. The DNA extracts were stored at -20°C until use.
Amplification and sequencing of the 16S rRNA gene and taxonomic identification of the bacterial cultures
The 16S rRNA gene fragments were amplified using the PCR universal bacterial primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’).11 The amplification was performed in Reaction mixture containing: 2.5 µL of PCR buffer 10 X, 0.8 mM of each dNTP (2.0 µL), 5.0 mM of MgCl (2.5 µL), 8.0 pmols of each primer, 1.25 U of Platinum® Taq DNA Polymerase (Invitrogen) (0.25 µL), 1 µL of the DNA of each culture pool and sterilized ultrapure water (Milli-Q) to a final volume of 25 µL. The conditions for the reaction were 95°C for five minutes, 30 cycles of 95°C for 30 seconds, 50°C for 30 seconds and 72°C for 90 seconds and 72°C for seven minutes. The partial sequencing of the 16S rRNA gene was executed in an ABI 3730xl DNA Analyzer capillary sequencer (Applied Biosystems, CA, USA). The 16S rRNA gene sequences were edited using Phred/Phrap.12 Taxonomy was assigned to the sequences using the Ribosomal Database Project Wang, et al.13 web-based taxonomy assignment tool version 2.6 against the RDP 16S rRNA training set 9.14
Statistical analysis
Spearman’s rank correlation coefficients were calculated between the relative abundance of bacterial phyla and physicochemical wastewater and water factors using the “multtest” package15 in R (R CoreTeam).16 P-values were corrected for multiple testing, using the false discovery rate controlling procedure.17 These coefficients were used for construction of bacteria-bacteria-physicochemical factors associations’ network. Cut-off of coefficient r >0.75 and significant positive and negative correlations (P values <0.07) were considered. The topology of the network was made based on average path length (parameter Network Interpretation: directed) Brandes18 and modularity (parameter randomize) Blondel et al.19 in order to determine the number of nodes in each network. Gephi open-source software20 was applied for analysis and visualization of the network.
Wastewater and water physicochemical characteristics
The analysis of wastewater and water physicochemical factors revealed lower DO concentration for the wastewater sample collected from the input point of the raw effluent for biological treatment (site 1) in comparison to the others samples. In turn, the wastewater sample collected from the output point of the effluent from stabilization pond (site 2) showed higher DO concentration than water samples collected from Jaguari River at the draw point of water for wastewater treatment (site 3) and Atibaia River, upstream the disposal of the effluent (site 4). BOD and QOD were also higher in site 1 than in any other site. Results of the physicochemical analysis are shown in Table 1.
Parameters |
Sampling Sites |
|||
Site 1 |
Site 2 |
Site 3 |
Site 4 |
|
pH |
7.81§ ±0.39 |
8.2±0.26 |
7.55±0.22 |
7.76±0.17 |
Conductivity (µS/cm) |
2303.78±525.86 |
2188.35±555.94 |
87.18 ± 21.23 |
245.96±70.16 |
Temperature (°C) |
30±2.76 |
26±3.13 |
22 ± 3.07 |
22±3.07 |
Transparency (cm) |
- |
- |
- |
53±25.2 |
Settleable solids (mL/L) |
1.07±1.88 |
<0.41± - |
0.12±0.04 |
0.61±0.71 |
Suspended soluble solids (g/L) |
17021±0.36 |
14209±0.46 |
0.1663±0.09 |
0.2806±0.1 |
Volatile matter (g/L) |
0.4611±0.24 |
0.3707±0.28 |
0.1073±0.08 |
0.1079±0.05 |
Remaining fixing material (g/L) |
1.26±0.38 |
1.05 ± 0.53 |
0.06±0.07 |
0.17±0.1 |
Apparent color (Ptco) |
1251±605.95 |
196±145.74 |
160±198.45 |
214±272.66 |
Turbidity (NTU) |
82.12±25.85 |
18.22±17.65 |
22.74±19.48 |
29.86±25.63 |
Dissolved Oxygen (mg/L) |
2.42±1.9 |
7.08±1.67 |
7.6±1.7 |
7.27±1.71 |
Chemical oxygen demand (mg/L) |
1102.04±665.39 |
137.47±52.42 |
13.77±9.93 |
21.44±14.36 |
Biochemical oxygen demand (mg/O₂/L) |
675.2±328.77 |
27.69±14.33 |
7.97±3.71 |
15.97±12.34 |
Chloride (mg/L) |
603.02±164.58 |
542.92±152.33 |
6.61±2.7 |
16.99±5.98 |
CaCO₃ acidity (mg/L) |
7.54±5.57 |
0±0 |
3.77±0.44 |
3.88±0 |
CO₃ alkalinity (mg/L) |
12.3±23.06 |
24.4±10.4 |
0±0 |
0±0 |
HCO₃ alkalinity (mg/L) |
275.69±47.47 |
188.29±30.64 |
27.68±4.09 |
38.09±7.36 |
Ammonia (mg NH₃/L) |
38.33 ± 14.64 |
1.9 ± 2.75 |
1.09 ± 0.45 |
2.15±1.47 |
Nitrite (mg/ NO₂/L) |
0.0371±0.01 |
0.2363±0.14 |
0.1387±0.08 |
0.2242±0.05 |
Nitrate (mg NO⁻³/L) |
115294±3.9 |
50916±2.95 |
16045±0.73 |
23848±1.78 |
Sulfide (µg/L⁻¹ S²⁻) |
175.5 ± 90.31 |
9.09 ± 6.13 |
8.61 ± 11.38 |
6.89±6.52 |
Cyanide (mg/L) |
3895±1.2 |
0.002±0 |
0.003±0 |
0.002±0 |
Oil and grease (mg/L) |
- |
1.1±0.63 |
- |
1.34±1.26 |
Phenols |
- |
0.84±0.68 |
- |
1720±1.27 |
Table 1 Physicochemical Characteristics of the Wastewater and Water Samples from the Different Sampling Sites.
§-Average for each of 18 sample replicates.
Standard deviation of the average for each of 18 sample replicates.
(-) Less than the detection limit.
Taxonomic identification of the bacterial isolated
Twelve different families of bacteria were identified based on the partial sequencing of the 16S rRNA gene of the bacterial cultures recovered from the four sampling sites. Members of Enterobacteriaceae, Paenibacillaceae, Pseudomonadaceae and Bacillaceae were ubiquitous in all samples, except for members of Bacillaceae, which were not recovered from site 2. These bacterial families revealed dominance over the other identified bacterial families, with highest relative abundance in each of four sites (Table 2). The result of the taxonomic identification of the 16S rRNA gene partial sequences of each of the bacterial isolated is shown in Supplementary Table 1 and Supplementary Table 2. The high abundance accounted for Enterobacteriaceae, Paenibacillaceae, Pseudomonadaceae and Bacillaceae may be a consequence of the laboratory methods used for cultivation and isolation of the bacterial cultures. The culture medium and incubation conditions at the lab may have favored the growth and isolation of members belonging to these families over recalcitrant members of the bacterial community.
Sampling Site |
Bacterial Family |
Relative Abundance |
Raw effuent (Site 1) |
Enterobacteriaceae |
63.64 |
Paenibacillaceae |
18.18 |
|
Pseudomonadaceae |
9.09 |
|
Carnobacteriaceae |
9.09 |
|
Output of the stabilization pond (Site 2) |
Enterobacteriaceae |
39.13 |
Bacillaceae |
21.73 |
|
Ferrimonadaceae |
4.35 |
|
Paenibacillaceae |
4.35 |
|
Aeromonadaceae |
4.35 |
|
Halomonadaceae |
4.35 |
|
Pseudomonadaceae |
8.69 |
|
Chitinophagaceae |
4.35 |
|
Flavobacteriaceae |
4.35 |
|
Oceanospirillaceae |
4.35 |
|
Jaguari River (Site 3) |
Enterobacteriaceae |
66.68 |
Bacillaceae |
9.52 |
|
Pseudomonadaceae |
9.52 |
|
Aeromonadaceae |
9.52 |
|
Paenibacillaceae |
4.76 |
|
Upstream the effluent discharge (Site 4) |
Enterobacteriaceae |
43.75 |
Synergistaceae |
6.25 |
|
Paenibacillaceae |
18.75 |
|
Bacillaceae |
25 |
|
Pseudomonadaceae |
6.25 |
Table 2 Relative Abundance of the Identified Bacterial Families in the Four Different Sampling Sites.
Co-occurrence pattern and theoretical relationships between bacterial families and wastewater and water physicochemical characteristics
The correlation level between bacterial families and physical and chemical parameters analyzed in this research was verified by Spearman’s rank correlation analysis and an artificial network of inter-relations between the two datasets was generated to illustrate it (Figure 2). The network highlights the relationships established by the families Aeromonadaceae and Carnobacteriaceae, which are represented by the biggest circles and therefore, are the ones that establish the highest number of theoretical relationships with the considered parameters and/or with other bacterial families. In addition, it was verified that both Aeromonadaceae and Carnobacteriaceae families establish relationships with three rather important parameters of water quality: DO, BOD and QOD, thereby, Aeromonadaceae is positively related (positive relationships are represented by thick connection lines) to DO and negatively related (negative relationships are represented by thin connection lines) to BOD and QOD. An opposite pattern was observed for Carnobacteriaceae family, which establishes negative relationship with DO and positive relationships with both BOD and QOD.

Figure 2 Artificial network of inter-relations between bacterial families and physical and chemical parameters from the analyzed effluent and water samples. The circumferences’ size indicates the quantity of correlations established by each of the considered physical and chemical parameters or bacterial family, e.g., the bigger the circumference the higher the number of theoretical relationships. The thickness of lines indicates if the relationships are positive or negative, so that thicker lines represent positive relationships and thinner lines, negative relationships.
It was observed that the families Bacillaceae, Enterobacteriaceae, Paenibacillaceae and Pseudomonadaceae were dominant in the analyzed samples, which may be due to the laboratory conditions of cultivation and isolation of the bacterial cultures, such as incubation temperature (36°C), which is known to be one of the major attributes that governs abundance and distribution of microorganisms. With the exception of Bacillaceae the other three previously mentioned bacterial families have mesophilic members,21-23 which means that these organisms develop best under median temperature and possess optimum temperature growth between 20 and 45°C. In a study about thermal water bacteria in India, Sen et al.24 noticed predominance of individuals coming from different groups; among them were Enterobacteriaceae, Paenibacillaceae, Bacillaceae and Pseudomonadaceae, the same families that were predominant in the analyzed samples of the present study. Enterobacteriaceae, Paenibacillaceae and Bacillaceae are facultative anaerobic bacteria families, being able to obtain energy in the absence of oxygen. This characteristic makes it easier for the members of these families to develop in laboratory conditions. Even though Pseudomonadaceae is a family of strict aerobic organisms it doesn’t have specific nutritive requirement, which makes it naturally ubiquitous and susceptible to development in laboratory culture media.
The inter-relations network for bacterial families and physical and chemical parameters sampled at different sampling sites in this research point theoretical relations established by the bacterial families Carnobacteriaceae and Aeromonadaceae with such parameters. Carnobacteriaceae are gram-positive, facultative anaerobic, rod-shaped bacteria which are generally associated to environments with decomposing organic matter.25 In this study members of the Carnobacteriaceae family were identified only in the raw effluent (site 1). Kim et al.26 showed members of Carnobacteriaceae family associated with tannery influent (raw effluent) generated in leather processing. Influent derived from both oil refining and tannery presents high load of organic compounds, besides many toxic agents. The theoretical relationship between members of the Carnobacteriaceae family and the presence of organic matter was pointed by the inter-relation network by showing the negative relationship between members of this family and DO levels and the positive relationships with both BOD and QOD. Such theoretical relationships suggest that Carnobacteriaceae family members may be associated with environments containing low dissolved oxygen rates and high biochemical and chemical oxygen demand rates, in which high amounts of decomposing organic matter are usually found.
On the other hand the Aeromonadaceae family is composed by gram-negative, facultative anaerobic, rod-shaped bacteria which can be found in many different types of aquatic ecosystems such as fresh water, seawater, brackish water and both clean and polluted water, moreover some members from this family occur exclusively in good quality water.27 Besides that, most of the bacteria belonging to this family are sensitive to the presence of chlorine and to very low concentrations of copper,27 both substances are present in the oil refining effluent. In the present study the Aeromonadaceae family was found in only two of the four sampling sites: in the output of the stabilization pond (site 2) and in the Jaguari River (site 3), both sites present higher dissolved oxygen average rates than the one found in raw effluent (site 1). Although the sampling site located upstream the effluent disposal (site 4) also presented higher dissolved oxygen rate than site 1, the tolerance of Aeromonadaceae to elevated levels of phenols may have affected the growth of members of this bacterial family in this location.28 The inter-relation network points theoretical relationships between Aeromonadaceae and DO (positive interaction) and BOD and QOD (negative interactions), which indicates members of Aeromonadaceae family can be present in systems full of oxygen and moderated amounts of organic matter. Aquatic environments with high concentrations of organic matter end up consuming the dissolved oxygen through chemical oxidation and, particularly, biochemical by the respiration of decomposing microorganisms. If the amount of organic material found in a water body exceeds its auto depuration capacity (capability of promoting the stabilization of a given polluting load) the dissolved oxygen rates drastically decrease, perhaps reaching zero and compromise the lives of the organisms inhabiting the water body. Therefore, the dissolved oxygen rates, the biochemical oxygen demand and the chemical oxygen demand represent excellent parameters for the evaluation of the level of organic pollution in lotic environments.29,30
Taken together, our findings demonstrate that differential occurrence of heterotrophic bacteria to specific wastewater and water physicochemical characteristics can help to reveal what drives their population changes in oil refinery wastewater treatment and adjacent water bodies and open the possibilities to explore Carnobacteriaceae and Aeromonadaceae families as potential indicators of oil refining effluent pollution and non-polluted or recovered environments, respectively.
This study was supported by a grant from Fundação para o Desenvolvimento da UNESP (FUNDUNESP) and Agência Nacional do Petróleo (ANP PRH-05). We wish to thank Valdenilson José Alves de Oliveira for his technical support during the wastewater and water sampling and Sibeli de Carli for her technical laboratory support.
The author declares no conflict of interest.

©2017 Becker, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.