International Journal of
eISSN: 2470-9980


Research Article Volume 2 Issue 2
1Islamic Azad University, Iran
2Department of Biology, Islamic Azad University, Iran
3Iranian Academic Center for Education, Iran
Correspondence: Mohammad Salehi, Medical Diagnostic Laboratory of ACECR-Razavi Khorasan, Ghavami Lane, Imam Khomeini Street, Neyshabour, Iran, Tel +98-935 842 8423, Fax +98-51-43337042
Received: January 30, 2016 | Published: April 7, 2016
Citation: Najafi S, Nazemi A, Salehi M (2016) The Survey of Association of HLA-B*51 and-B*52 Alleles with HSV, CMV by Specific Sequence Primer-PCR. Int J Vaccines Vaccin 2(1): 00025. DOI: 10.15406/ijvv.2016.02.00025
Background and Aim: Major Histocompatibility Complex (MHC) is the most polymorphic system in the genome of different species. In human beings, it named human leukocyte antigen (HLA). The association between genetics, particularly the genes of the MHC and many Infection diseases especially viral infection has been known. The aim of this study is survey of association of HLA-B*51 and -B*52 alleles with HSV, CMV infections by Specific Sequence primer-PCR (SSP-PCR).
Materials and Methods: Eighty eight people referred to the Pathobiology Laboratory of Rasht city, Iran who suspected to HSV and CMV infections. The Real-Time PCR was used for detection of HSV and CMV infections. HLA typing was performed by using SSP-PCR as a precise method.
Results: In this study, 6 of 88 samples were positive for HSV infection (6.81%) and 9 for CMV infection (10.22%). Although HLA typing indicate the 2.53% (2/79) of healthy individuals expressed the HLA-B*51 and -B*52 alleles. In contrary there is no expression of alleles in patients. (0/6).
Conclusions: Results of this research showed that there is no significant relation between the HLA-B*51 and -B*52 alleles and being resistant or catching the HSV or CMV infections (P > 0.05).
Keywords: HLA-B*51 and -B*52, CMV, HSV-1 & HSV-2, Specific sequence primer-PCR
MHC, Major Histocompatibility Complex; HLA, Human Leukocyte Antigen; CMV, Cytomegalovirus; HSV, Herpes Simplex Virus; CSF, Cerebrospinal Fluid; SSP-PCR, Specific Sequence Primer-PCR; BD, Behçet’s disease
The herpes family of viruses includes a number of major human pathogens such as herpes simplex virus (HSV) and human cytomegalovirus (CMV) that cause a wide range of diseases, including herpes, genital herpes and cytomegalic inclusion.1
A variety of human diseases have recently been found to occur more frequently in individuals possessing certain antigens called histocompatibility or tissue antigens.2 The major histocompatibility complex (MHC) is a set of cell surface proteins essential for acquired immune system to recognize foreign molecules in vertebrates, which in turn determines histocompatibility. The MHC region occurs on chromosome 6 and contains 240 genes. In humans, the MHC is also called the human leukocyte antigen (HLA). Each human cell expresses six MHC class I alleles (one HLA-A, -B, and -C allele from each parent), six to eight MHC class II alleles (one HLA-DP and -DQ, and one or two HLA-DR from each parent, and combinations of these) and MHC Class III molecules that have physiologic roles.3
B51 is a split antigen of the broad antigen B5, and is a sister serotype of B52. There are a large number of alleles within the B*51 allele group. B51 is associated with several diseases.4
Clinical trials, epidemiological and demography studies reflects the special relationship between certain diseases and a specific antigen of HLA. Although the nature of this relationship is unclear, but generally such that diseases thought to be associated with certain genetic background.5 Also, exposures to some viruses invoke a variety of genetically-controlled immunological responses. These include the activation of T, B and natural killer (NK) cells, as well as the production of antibodies and a range of cytokines, which together can be either protection or detrimental to individuals exposed to viral infections.6
Some studies showed the present of some specific HLA alleles can make the person more resistant or more sensitive than the disease. Until now, the relationship between expression of HLA genes with resistance or sensitivity of some disease such as Behqet’s syndrome, myasthenia gravis, multiple sclerosis, HIV, severe malaria, Leishmaniasis and Toxoplasmosis have been demonstrated and also the role of some HLA alleles including of HLA-B*51, HLA-B*18 and HLA-A*28 in establishment of infection have been reported.7‒13 Also, it can be mentioned to high frequency of HLA-B*27 in spondylo arthropathies, HLA-DR3 and HLA-DR4 in diabetes and HLA-DR4 in rheumatoid arthritis.14‒16
The cause of some diseases like Behçet’s (BD) remains unknown, but epidemiologic findings suggest that an autoimmune process is triggered by an environmental agent in a genetically predisposed individual.17,18 Among them, several viral agents, including herpes simplex virus-1 and cytomegalovirus may also have some role.19,20 Herpes simplex virus-1 (HSV-1) is currently the most common virus associated with BD. HSV DNA and serum antibodies against the virus have been found in a higher proportion of patients with BD than in controls.20 It has long been known that BD is associated with the HLA-B5 allele21 and, more specifically, with HLA-B51.22 So, it can be assumed that there is an association between HLA-B5 alleles and HSV or CMV infections.
Also, HLA genes are a property which is not only central to their pivotal role in the recognition of self from non-self antigens, but also makes the antigens encoded by these genes important in organ donation and recipients/donor compatibility. HLA typing also provides a useful tool for studies of human population dynamics, migration, and colonization.23 Anthropological studies, and determination of HLA allele and haplotype frequencies in different ethnic groups, have been found to be a valuable tool in population genetic analyses and the study of genetic relationships. Amirzargar et al.,24 have reported DRB1*11 as the most frequent allele among People of Fars Ethnicity Living in Iran.25 As well as, there are significant differences in the prevalence of some viral infections among different regions of the world that seem to be due to the differences in the frequency of alleles of the HLA.26
Futohi et al.,27 have investigated the relationship between 59 HLA alleles and the CMV infection, in transplant recipients, after kidney transplantation. Findings showed that deceased donor renal transplantation and the presence of HLA-B44 can make the kidney recipient susceptible to CMV infection after kidney transplantation; on the other hand, the presence of HLA-B8 can have a protective effect.27
A study conducted in 1981 by Volker were studied the distribution of antigens HLA-A, B, C in 115 patients with recurrent herpes keratitis and 123 healthy individuals as controls. The results showed HLA-A*3 significantly (p <0.042) was associated with risk of herpes viruses.28
The association of the prevalence of HSV-1 & HSV-2 and CMV infections and diseases with the frequency of human leukocyte antigens (HLA) alleles is not fully elucidated. Some studies point to association trends between the frequency of some common HLA alleles and the high/low prevalence of HSV-1 & HSV-2. While, few studies have reported association of HLA alleles with CMV infection.10,26,29
In this study, we investigated the association of HLA-B*51 and-B*52 alleles with HSV and CMV by Specific Sequence primer-PCR. In fact, we intent to find an association between the frequency of these alleles and the sensitivity or resistance to these diseases.
Collecting data and samples
This cross-sectional study (case-control) was performed for molecular detection of HLA-B*51 and-B*52 alleles on 88 patient (age range 22 to 35years) who suffering of HSV or CMV disease. In order to HSV or CMV testing, the plasma and cerebrospinal fluid (CSF) were gathered from 2014 to 2015 in patient who referred to Pathobiology laboratories of Rasht city.
The samples were retained in -20 °c by the time of examination. In order to HLA test, 5ml of whole blood is poured into lithium heparin tubes and immediately were stored at refrigerator temperature. Since HLA testing is only done on fresh samples, so the test was performed on the same day. Approval for this study was obtained from University of Medical Sciences of Rasht Branch. Informed consent was obtained from patients.
DNA extraction and testing
DNA extraction was done for all samples (Dynabio DNA Extraction Mini Kit, IRAN). Real Time PCR method was used for detection CMV and HSV in serum as a reliable and more sensitive method.30 The Kit can be used for both qualitative and quantitative method. we aimed to identify the presence of the virus, That is why, the qualitative method was used.
The sequence of primers that were used in current study was confidential by relevant company duo to avoiding any abuse. In order HLA typing, we detected the HLA-B*51 and-B*52 alleles by use of PCR-SSP method (PZP Kit, IRAN). DNA-based HLA typing methods have much higher sensitivity, accuracy and diagnostic strength than serological methods.31 The positive DNA control supplied in the kit is used as a control to test the alleles for SSP-PCR and Real Time PCR.
Statistical analysis
Data were analyzed with SPSS version 20 (IBM SPSS Statistics for Windows, Version 20, Armonk, NY, IBM Corp.), the chi-square test. p-values <0.05 were considered statistically significant.
The mean age of participants is 29.66±8.48years old. The accuracy of DNA extraction has confirmed. According to Real Time PCR results, 9 cases of CMV infection and 6 cases of HSV infection were positive (Table 1).
Type of Virus |
Seropositivity Case (%) |
Seronagativity Case (%) |
CMV |
9 (10.22) |
79 (89.77) |
HSV |
6 (6.81) |
82 (93.18) |
Total |
15 (17.04) |
73 (82.95) |
Table 1 The results of the risk of viral infections in patients with suspected CMV and HSV infection
The result of PCR for HLA-B*51 and-B*52 alleles
The result of SSP-PCR for HLA-B*51 and-B*52 alleles have determined by amplification of a 455 base pair length nucleotide. Results also demonstrated among 73 negative samples, only 2 people had the HLA-B* 51 and-B* 52 alleles. No one of 15 positive samples had the HLA-B* 51 and -B* 52 alleles (Figure 1-3)( Table 2).
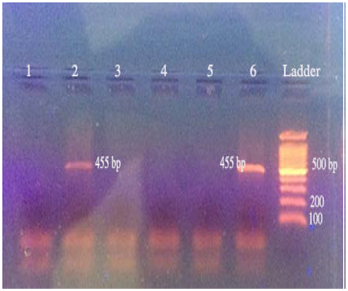
Figure 1 The results of electrophoresis of the reaction product of HLA-B* 51 and-B* 52 alleles on some samples.
Rows 1 to 4 is participant samples, row 5 is negative control(nuclease-free water), row 6 is positive control and the last row is DNA molecular weight marker (PZP, Iran).
CMV |
Negative |
Positive |
Total |
Odd Ratio |
95% Confidence Interval |
p-value |
Female |
52 |
0 |
52 |
1.333 |
1.104-1.610 |
<0.0001 |
Male |
27 |
9 |
36 |
|||
Total |
79 |
9 |
88 |
|
|
|
Table 2 The relation of CMV infection with sex
The results of statistical analysis rejected the association between HLA-B* 51-and-B* 52 allele and it’s relation with sensitivity to CMV infection (p = 0.734) and HSV infection (p = 0.786).
HLA-B*51 and - B*52 alleles were expressed only in 2 of 73 without infection samples and statistical analysis showed no relationship between CMV/HSV infections and susceptibility or resistance to these infections (Table 3-5).
CMV |
Negative |
Positive |
Total |
Odd ratio |
95% confidence interval |
p-value |
Absence alleles of HLA-B*51 and-B*52 |
78 |
9 |
87 |
0.897 |
0.835-0.963 |
0.734 |
Presence alleles of HLA-B*51 and-B*52 |
1 |
0 |
1 |
|||
Total |
79 |
9 |
88 |
|
|
|
Table 3 The relation of CMV infection with absence or presence of HLA-B*51 and-B*52
HSV |
Negative |
Positive |
Total |
Odd Ratio |
95% Confidence Interval |
p-Value |
Female |
52 |
0 |
52 |
1.2 |
1.037-1.389 |
0.002 |
Male |
30 |
6 |
36 |
|||
Total |
82 |
6 |
88 |
|
|
|
Table 4 The relation of HSV infection with sex
HSV |
Negative |
Positive |
Total |
Odd ratio |
95% confidence interval |
p-value |
Absence alleles of HLA-B*51 and-B*52 |
81 |
6 |
87 |
0.931 |
0.879-0.986 |
0.786 |
Presence alleles of HLA-B*51 and-B*52 |
1 |
0 |
1 |
|||
Total |
82 |
6 |
88 |
|
|
|
Table 5 The relation of HSV infection with absence or presence of HLA-B*51 and-B*52
Samandary et al.,26 showed the high frequency of HLA-A*24, HLA-B*27, HLA-B*53 and HLA-B*58 is associated with occurrence of HSV-1 & HSV-2. In contrast, the low frequency of HLA-B*44 is associated with HSV1-2 in Spain.26
In the study of Paya et al.,30 found no association between cytomegalovirus infection and HLA-B antigens 30. The result of this study was in agreement with current study.
Zimmerman et al.,32 investigated the HLA antigens in 46 people with recurrent herpes simplex infection of the cornea and compared with the control group. The results showed that HLA-B5 antigen significantly more common in patients in compare to control group32 Also, colin et al.,33 HLA antigens in 80 patients with recurrent herpes eye infection compared to the control group. Based on the results, HLA-B5 in the patient group was significantly more common.
Frjadyan et al.,24 reported, HLA-B*35 allele is the most abundant locus of HLA-B (8/28%) in the Iranian population who has the lowest incidence of herpes infections.24
Stephens et al.,34 were investigated the HLA allele associations with secondary dengue virus infections and it’s correlation with disease severity and infecting viral serotype in 263 ethnic Thais. The result showed HLA-B*51 and HLA-B*52 Was associated with development of DHF and DF disease in patient with secondary dengue virus infections respectively.34
Zhang et al.,10 reported multilayered defense in HLA-B51-associated HIV viral control. They propose that patients with HLA-B51 benefit from having multiple layers of effective defense against the development of immune escape mutations.10
Differences between the results of current study and other studies around the world may be due to many factors including geographic distribution, different races, different viruses and HLA alleles.
Identification of high risk population by further studies and appropriate methods can effective in health planning, vaccination and other prevention strategies. So, we proposed in order to more precise investigation, researchers do this study with more samples and other HLA’s alleles.
By regard of the results of this study the association between HLA-B* 51-and-B* 52 allele and it’s relation with sensitivity to CMV infection (p = 0.734) and HSV infection (p = 0.786) are rejected and there is no significant relation between HLA-B*51 and -B*52 allele and sensitivity or resistance to these infections.
This article is the result of the thesis of Somaye Najafi that recorded by No. 15930507932007 for receive a master’s degree in Biology (Microbiology) from the Islamic Azad University. The authors are grateful from the personnel of the Islamic Azad University.
Author declares there are no conflicts of interest.
None.

©2016 Najafi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.