International Journal of
eISSN: 2573-2889


Research Article Volume 3 Issue 5
Medical Laboratory Technology Department, Taibah University, Saudi Arabia
Correspondence: Samia Mahdi Ahmed Mohammed, Medical Laboratory Technology Department, Taibah University, Saudi Arabia
Received: June 18, 2018 | Published: October 9, 2018
Citation: Mohammed SMA, Algarni BA, Almugari BF, et al. Screening of ass1 gene in two Saudi families from al-madinah al-monawarah with citrullinemia disorder. Int J Mol Biol Open Access. 2018;3(5):222-233. DOI: 10.15406/ijmboa.2018.03.00081
Background: Citrullinemia type 1 is an inherited autosomal recessive disease and a member of Urea Cycle Disorders (UCD),1 characterized by accumulation of ammonia in the blood as a result of defect in enzyme called “argininosuccinate synthetase” (ASAS), which is responsible for converting citrulline to arginine. Signs and symptoms appear after birth, if untreated CTLN1 may progress to coma or death.
Purpose: To Screen ASS1 gene in two Saudi families with citrullinemia disorder (one affected family and the other is a carrier) from AL- Madinah AL-Monawarah. Define mutations may help in treatment and cure progress for patients.
Methods: Genetic analysis using sequencing technology was carried out to detect mutations in ASS1 gene.
Results: Two Mutations were detected. First family affected and unaffected members (III:3, II:2) have a homozygous missense variant in exon 7 (c.501 C>T, p.166 His>His). While in second family affected member (III:1) has a homozygous splice site mutation in exon 5 (c.364-2 A>G) and unaffected member (II:2) has a heterozygous mutation in exon 5 (c.371 A>T) of ASS1 gene.
Conclusion: Missense and splice site mutations were found in both affected and carrier members, homozygous and heterozygous respectively. The splice site region is important because mutations in these areas may lead to entire exon being spliced out of the mRNA. A heterozygous mutation makes individual a carrier of the disease.
Research Objectives
To compare ASS1 sequence in affected individuals and his/her unaffected family members.
Citrullinemia is life-threatening disease, it occurs in 1:57,000 births and usually becomes evident in the first few days of life. It belongs to a small number of conditions called urea cycle disorders (UCD)1 and it is characterized by accumulation of ammonia in the blood because of defect in enzyme called argininosuccinate synthetase (ASAS), which is either being missing or not working properly. ASAS is responsible for converting citrulline to arginine in urea cycle. Accumulated citrulline and ammonia impair the organ's antioxidant capacity.2
In citrullinemia condition, argininosuccinate synthetase enzyme is absent or insufficient due to mutation in Argininosuccinic acid synthetase (ASS1).3
Increasing knowledge of mutations that leads to ASAS abnormalities would help in better treatment and cure. This research is trying to define more mutations in ASS1 gene.
Citrullinemia definition
Ammonia and other toxic substances can be accumulated in the blood because of an inherited autosomal recessive disorder (Figure 1.1 & Figure 1.2) (Table 1.1) called citrullinemia. Citrullinemia belongs to a family of genetic diseases named urea cycle disorders. Mutations in the ASS1 and solute carrier family 25, member 13 (citrin) SLC25A13 genes cause citrullinemia. The urea cycle is a series of chemical reactions that work at excess nitrogen generated when protein is used by the body in liver cells. The excess nitrogen is used to make a compound called urea, which is excreted in urine.4,5
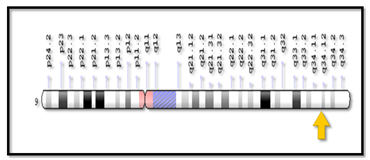
Figure 1.2 Cytogenetic and molecular location of argininosuccinic acid synthetase (ass) gene (genetics home reference, 2006).
1.2.3 Mutations Found in |
Protein |
mRNA |
DNA |
Nonsense |
|
|
|
Int 0 |
c.-4C4T |
||
Ex 7 |
p.G156X |
c.459_466del |
|
p.N158X |
c.450_451delCT |
||
Int 11 |
r.773_774ins47 |
c.773149C4T |
|
Ex 12 |
p.G275X |
c.823G4T |
|
Ex 12 |
p.R279X |
c.835C4T |
|
Ex 13 |
p.Q311X |
c.931C4T |
|
Ex 14 |
p.R344X |
c.1030C4T |
|
Ex 15 |
p.Q380X |
c.1138C4T |
|
Int 15 |
p.Q401X |
c.1194-21_1213del |
|
Splice site |
|||
Int 4 |
c.17411G4T |
||
Int 6 |
c.421-2A4G |
||
Int 7 |
r.421_495del |
c.49511G4T |
|
Int 11 |
c.77311delG |
||
Int 11 |
c.77312_77313insT |
||
Int 12 |
c.83811G4A |
||
Int 13 |
r.839_970del |
c.97015G4A |
|
Int 14/ Ex 15 |
r.1127_1128ins67 |
c.1127-9_1185dup67 |
|
Int 15 |
r.Ex16del7 |
c.1194-1G4C Turkey |
|
Int 15 |
p.L399AfsX409 |
r.1193_1194ins37 |
|
Deletion |
|||
Ex 5 |
p.R100PfsX139 |
c.299delG |
|
Int 7/Ex 8 |
c.496-21_515del |
||
Ex 11/Ex 12 |
r.689_838del |
||
Ex 13 |
p.E298RfsX315 |
c.892delG |
|
Ex 13 |
p.A318LfsX375 |
c.952delG |
|
Missense |
|||
Ex 3 |
p.G14S |
c.40G4A |
|
p.S18L |
c.53C4T |
||
p.C19R |
c.55T4C |
||
Ex 4 |
p.Q40L |
c.119A4T |
|
Ex 5 |
p.V69A |
c.206T4C |
|
p.S79P |
c.235T4C T |
||
p.R86C |
c.256C4T |
||
p.R86H |
c.257G4A |
||
p.R95S |
c.285G4T |
||
p.P96S |
c.286C4T |
||
p.P96H |
c.287C4A |
||
p.R108L |
c.323G4T |
||
p.G117S |
c.349G4A |
||
p.G117D |
c.350G4A |
||
p.A118T |
c.352G4A |
||
p.T119I |
c.356C4T |
||
Ex 6 |
p.D124N |
c.370G4A |
|
p.R127W |
c.379C4T |
||
p.R127Q |
c.380G4A |
||
Ex 7 |
p.R157C |
c.469C4T |
|
p.R157H |
c.470G4A |
||
p.L160P |
c.479T4C |
||
Ex 8 |
p.W179R |
c.535T4C |
|
p.S180N |
c.539G4A |
||
p.Y190D |
c.568T4G |
||
Ex 9 |
p.E191K |
c.571G4A |
|
p.E191Q |
c.571G4C |
||
p.A192V |
c.575C4T |
||
Ex 10 |
p.A202E |
c.605C4A |
|
p.L206P |
c.617T4C |
||
Ex 12 |
p.V263M |
c.787G4A |
|
p.R265C |
c.793C4T |
||
p.R265H |
c.794G4A |
||
p.V269M |
c.805G4A |
||
p.E270Q |
c.808G4C |
||
p.R272C |
c.814C4T |
||
p.K277T |
c.830A4C |
||
p.R279Q |
c.836G4A |
||
p.G280R |
c.838G4C |
||
Ex 13 |
p.E283K |
c.847G4A |
|
p.T284I |
c.851C4T |
||
p.Y291S |
c.872A4C |
||
p.D296G |
c.887A4G |
||
p.M302V |
c.904A4G |
||
p.R304W |
c.910C4T |
||
p.R307C |
c.919C4T |
||
p.K310Q |
c.928A4C |
||
p.K310R |
c.929A4G |
||
p.G324S |
c.970G4A |
||
Ex 14 |
p.G324V |
c.971G4T |
|
p.S341F |
c.1022C4T |
||
p.V345G |
c.1034T4G |
||
p.G347R |
c.1039G4C |
||
p.Y359D |
c.1075T4G |
||
p.G362V |
c.1085G4T |
||
p.R363W |
c.1087C4T |
||
p.R363G |
c.1087C4G |
||
p.R363L |
c.1088G4T |
||
p.R363Q |
c.1088G4A |
||
Ex 15 |
p.T389I |
c.1166C4T |
|
|
p.G390R |
|
c.1168G4A |
Table 1.1 Mutations of the ASS1 Gene10
Citrullinemia types
1- CTLN2 is a disorder caused by the lack of SLC25A13 gene, which is responsible for making citrin protein. Citrin is a helpful protein because it has four functions, which are build up and breakdown of sugars, proteins synthesis, transportation of urea cycle molecules and it takes part in the production of nucleotides. Any mutations in SLC25A13 gene blocks the functional citrin, it leads to inhibition of urea cycle and production of nucleotides and proteins which causes accumulation of ammonia and other toxic substances in adult-onset.6
It is adult-onset (11 to 79 years old) usually from adolescence to adulthood. it’s effect on the nervous system suddenly and cause lack of understanding, amnesia, dysfunctional behavior, coma due to increase of ammonia in blood and leads to death because of brain edema.7
Diagnosis of CTLN2 depends on two tests, which are biochemical test and mutational study. The result of the biochemical test is hyperammonemia, the amino acids concentration will be increased with the increasing of concentration of citrulline and arginine can be showing in blood profile. While the confirmatory test is done by mutational study of the SLC25A13 gene, allowing genetics counseling.8
CTLN2 is a life-threatening disorder because of untreated cerebral edema. The only effective treatment is liver transplantations.9
2- Citrullinemia type 1 (CTLN1) is caused by mutations in ASS1 gene, which provides instructions for making an enzyme called argininosuccinate synthase. It is responsible for the third step of Urea Cycle. This step combines two protein building blocks (amino acids), citrulline and aspartate, to form a molecule called argininosuccinic acid. A series of additional chemical reactions uses argininosuccinic acid to form urea.10
According to Genetics Home Reference: "Cytogenetic Location of ASS1: 9q34.11, which is the long (q) arm of chromosome 9 at position 34.11" (Figure 1.2).3
According to Genetics Home Reference: "Molecular Location of ASS1: base pairs 130,444,707 to 130,501,274 on chromosome 9" (Figure 1.2) (Table 1.1).3,10,11
The Prevalence of CTLN1 among the world has been estimated to occur in 1:57,000 births.12 Newborn screening programs found CTLN1 in different country:13
The prevalence of CTLN2 disorder is less common from CTLN1, High frequency of this disorder founded in Japanese population about 1:100,000-230,000. It also recorded in Middle East and East Asia.19
Citrullinemia signs and symptoms
CTLN1 symptoms appear soon after birth and caused by increase ammonia level in blood. CTLN1 takes place usually after protein feeding in the first two to three days of life and start with: Absence of appetite refusal to feed, emesis, doziness, lack of energy, irritability, abnormally rapid breathing. In addition, if ammonia level increased more without treatment it may cause low muscle tone (hypotonia). respiratory problems, uncontrolled body temperature, liver enlargement, lack of growth, swelling of the brain due to increasing of fluid, increased intracranial pressure, seizures, ankle clonus, with increased ammonia level patient could go to hyperammonemic coma resulting in Neurological development delay, intellectual disability, learning difficulties, if untreated it could cause death in few weeks after birth.20
Citrullinemia diagnosis
Diagnosis of citrullinemia can be confirmed by a detailed patient family history, identification of characteristic findings, and a variety of specialized tests. Excessive amounts of ammonia and citrulline in the blood strongly suggests the diagnosis of CTLN1. Molecular genetic testing for ASS1 gene mutations is available to confirm the diagnosis.21
Newborn Screening Results: Elevated citrulline is detected in dried blood spots on newborn screen by tandem mass spectroscopy (MS/MS).20
Neonatal presentation: Sign and symptoms classically occur within the first week of life while on a full protein diet:
Supportive laboratory findings
Molecular Genetic Testing: Molecular testing is helpful when the phenotype is unclear, when biochemical values are borderline, or to distinguish CTNL1 from CTNL2.10
ASS1 is the only gene associated with CTNL1. The majority of causative mutations in the gene are sequence variants, but large deletions have been reported, The Methods to detect mutations of ASS1 gene PCR (Polymerase chain reaction) followed by bidirectional sequencing of the entire coding region and intron/exon boundaries of the ASS1 gene.10
Citrullinemia treatment & management
The treatment of hyperammonemia22 should be applied immediately to prevent and avoid complications which associated with neurologic damage or even death with severe untreated hyperammonemia patients. Intellectual ability is inversely related to the length of time of infant with urea cycle disorder without treatment. Immediate treatment is extremely important in avoiding or minimizing the degree of complications.23
Materials
Ethical consideration: The study proposal was reviewed and ethically approved by the Scientific and the Ethical Committee (Institutional Review Board) at Taibah University and Institutional Review Board committee of health affairs in Madinah. Written informed consents were obtained from all individuals.
Study design: This is a familial study. Following informed consents, individuals with citrullinemia were enrolled in this study.
Study area: The study was conducted in Al-Madinah Al-Monawarah. Patients were recruited from Maternity Hospital. Genetic analysis including DNA extraction, PCR, and sequencing was performed in Center of Genetic and Inherited Diseases (CGID).
Study population: Two families from Al-Madinah Al-Monawarah were included in this study after being fully informed about the aims of the study. The results of the analysis (gene screening and sequencing), have been used for clinical diagnosis, and offered free of charge for all patients participating in the study.
First family was recruited from Al-Madinah Al-Monawarah. It is a three generations autosomal recessive CTLN1 family with 10 members with no history of any other genetic disease. In this family, 8 individuals are unaffected (I:1, I:2, I:3, I:4, II:1, II:2, III:1, III:2) and 2 individuals are affected (III:3, III:4) (Figure 2.1). Both affected individuals are females. Affected individual (III:3) is a 4 years old female diagnosed with CTLN1 at birth. Affected individual (III:4) is a 6 years old female diagnosed with CTLN1 at birth. Blood sample were collected from 1 affected (III:3) and 1 unaffected member (II:2) for deoxyribonucleic acid (DNA) extraction.

Figure 2.1 Pedigree of the first family from al-madinah al-monawarah. it is three generations autosomal recessive ctln1 family.
The second family was recruited from Al-Madinah Al-Monawarah. It is a three generations autosomal recessive CTLN1 family with 11 members with no family history of any other genetic diseases. In this family, 9 individuals are unaffected (I:1, I:2, I:3, I:4, II:1, II:2, III:3, III:4, III:5) and 2 individuals are affected (III:1, III:2) (Figure 2.2). Both affected individuals are males. The affected individuals (III:1) is a 4 years old male diagnosed with CTLN1 at birth. Affected individual (III:2) is a 7 years old male diagnosed with CTLN1 at birth. Blood samples were collected from 1 affected (III:1) and 1 unaffected member (II:2) for DNA extraction.

Figure 2.2 Pedigree of the second family from Al-Madinah Al-Monawarah. It is three generations autosomal recessive CTLN1 family.
Medical history: The individuals were interviewed by trained team, demographic and clinical data about the patients and their families was collected in a specially designed questionnaire (Appendix 1). Demographic data included age, sex, tribe, weight, height, and address. Clinical data included medical history of citrullinemia, type of treatment, type of diet, ammonia level measurement.
Blood collection: Blood samples (3ml) were collected from available participants in 4 ml EDTA (Ethylenediaminetetraacetic acid) tubes and stored at 4◦C.
Methods
DNA extraction and quantification: The blood collected in EDTA tubes was equilibrated to room temperature and extraction of DNA was carried out using QIAGEN (QIAamp® DNA) Mini Kit according to the supplier’s protocols. The DNA was quantified by Nanodrop spectrophotometer (MAESTRO) and diluted to 40ng/µl for PCR (Table 2.1 & table 2.2).
Patients |
DNA Purity |
DNA concentration |
III:3 |
1.9 |
22 |
II:2 |
2 |
20 |
Reference Values |
1.7 - 2 |
20 – 50 |
Table 2.1 DNA quality and concentration determined by nanodrop spectrophotometry for the first family
Primer name |
Sequence |
Temperature |
Product size |
ASS1_Ex2F |
CAGGAGACAAGGCTGTCC |
56.69 |
318 bps |
ASS1_Ex2R |
GAGCAGACAGGCTGACAAC |
56.96 |
318 bps |
ASS1_Ex3F |
ATGGTGTGAACTCAGGGC |
56.83 |
202 bps |
ASS1_Ex3R |
CAGGAGCATCCACCACTG |
58.67 |
202 bps |
ASS1_Ex4F |
GGGCTCTGTATGCCAGATG |
59.21 |
320 bps |
ASS1_Ex4R |
CTCATCTCCTTCCCACACC |
57.98 |
320 bps |
ASS1_Ex5F |
CTGTCCTTGTCCTCACGTC |
56.61 |
242 bps |
ASS1_Ex5R |
ATTCTGTGCCTGTCCTGTG |
57.1 |
242 bps |
ASS1_Ex6F |
CCTCACAACAGCATCCTCTC |
58.38 |
375 bps |
ASS1_Ex6R |
ACAGAGGCCACGTGTGTAG |
57.21 |
375 bps |
ASS1_Ex7F |
AGGGTCTTGTCTGAATGGG |
57.47 |
244 bps |
ASS1_Ex7R |
GTCCTTTGGAATGAGCCC |
57.98 |
244 bps |
ASS1_Ex8F |
ATGTTTCAGGCAGGTTGG |
56.97 |
231 bps |
ASS1_Ex8R |
AGCTGCTACCACCAAAGG |
56.38 |
231 bps |
ASS1_Ex9F |
GTAGGGTGTCCAGGGACTG |
57.9 |
264 bps |
ASS1_Ex9R |
TAGCTTCCAGGAATGCAGG |
58.96 |
264 bps |
ASS1_Ex10F |
ATCCATTTAAGGCGTTTCG |
57.71 |
280 bps |
ASS1_Ex10R |
CTCAGCCACAACCATTAGC |
56.83 |
280 bps |
ASS1_Ex11F |
GGTGACTCTGAGCCTTGC |
56.87 |
178 bps |
ASS1_Ex11R |
GATCTCTGGGTTTGCTGG |
56.58 |
178 bps |
ASS1_Ex12F |
GTCATTTGCTGACAGTTTGG |
56.72 |
265 bps |
ASS1_Ex12R |
GTACTTTGGGATCCCTTGTG |
56.96 |
265 bps |
ASS1_Ex13F |
CCCAGGTCTCCCTGTGTC |
58.97 |
308 bps |
ASS1_Ex13R |
TCAGGCACAGATGTCTTGAG |
57.44 |
308 bps |
ASS1_Ex14F |
CAGTCCTCCCTTCAAGCAG |
58.52 |
190 bps |
ASS1_Ex14R |
CAAAGCCTACCCTTTACACC |
56.4 |
190 bps |
ASS1_Ex15F |
ACCCAGTGTGTGTTGTTATTG |
55.89 |
181 bps |
ASS1_Ex15R |
TTATCACAACAATTAGCGCC |
56.42 |
181 bps |
Table 2.2 DNA quality and concentration determined by nanodrop spectrophotometry for the second family
Genomic DNA visualization: One percent (1%) of agarose gel was prepared by mixing 50 ml 1X-Tris-borate-EDTA (TBE) buffer and 0.5 g agarose. Solution was heated to boiling, followed by cooling and 3µl of Ethidium Bromide (EtBr) was added. Solution was poured to casting tray.
Three µl of the loading dye was mixed with 3µl of the sample (genomic DNA)23 and loaded into gel wells. Electrophoresis was performed at 120 voltages for 40 minutes. Gel was visualized by Ultraviolet (UV) in a gel documentation system (Gensys). Ladder size was 10bp (1000-100).
Primers designing: Primers were designed using Primer 3 software from University of California Santa Cruz (UCSC) genome browser (www.genome.ucsc) website. Primer sequences, melting temperature and size of amplified product of all exons of ASS1 gene is available in (Table 2.3). Exons 1 and 16 is non-coding sequences.25
Patients |
DNA Purity |
DNA concentration |
III:1 |
1.8 |
20 |
II:2 |
1.9 |
21 |
Reference value |
1.7 - 2 |
20 - 50 |
Table 2.3 Primer sequences melting temperature and size of amplified product of all exons of ASS1 gene).
Exon amplification
ASS1 gene was amplified by PCR. Amplified products were analyzed on 2% agarose and purified by QIAquick Purification kit (QIAGEN). Polymerase chain reaction was carried out in 50µl reaction volume containing 5ul of genomic DNA, 2.5ul of each primer (forward primer and reverse primer), 25ul of master mix (GO) and 15 µl distilled water. The thermal cycler condition used were 35 cycles, 95°C for 1 minute (denaturation), 57-60°C for 1 minute (annealing), 74°C for 1 minute (extension), and final extension of 72°C for 10 minutes. PCR was performed by Veriti 96 well thermal cycler (Applied Biosystems). Annealing temperature for exons 2,3,4,6,8,9,10,11,12,13,14 was 57◦C, 58◦C for exon 15, and 59◦C for exons 5 and 7.26–30
First purification of amplified products
Amplified products were purified using USB® ExoSAP-IT®PCR Product Cleanup Kit according to the supplier’s protocols.
Second amplification of purified product (second PCR)
One µl of DNA templates (from first purification) were mixed with 1µl 5X sequencing buffer, 6 µl distilled water (dH2O), 1µl of (forward or reverse) primer and 1µl of Big dye terminator. They were mixed together and then centrifuged; they were placed on thermal cycler machine for amplification.
The thermal cycler condition included 35 cycles, 95°C for 3 minutes, 95°C for 1 minute, 57°C for 30 seconds, 60°C for 1 minute and final extension of 60°C for 4 minutes. PCR was performed by Veriti 96 well thermal cycler (Applied Biosystems).
Second purification of amplified products
Amplified products were purified using Big Dye® X-TerminatorTM Purification Kit (Applied Biosystems) according to the supplier’s protocols.
Sequencing
After mixing the purified product by Mixmate, the supernatant was transferred to the sequencing plate for sequencing in the capillary electrophoresis and chromatograms were generated on ABI 3500 genetic analyzer (Applied Biosystems).
In the present study, two Saudi families with CTLN1 disease with no history of any other genetic disease were recruited from Al-Madinah Al-Monawarah have been investigated . DNA was extracted from all available individuals. Two blood samples collected from family 1(III:3 affected, II:2 unaffected) and two blood samples collected from family 2(III:1 affected, II:2 unaffected) (Figure 3.1). DNA extraction and Gel Electrophoresis was done on the samples with 1% agarose gel concentration.
DNA for four (III:3, II:2, III:1, II:2) individuals both affected and unaffected were amplified by PCR (Figure 3.2-3.4), purified and sequenced under standard conditions. Sequence analysis using reference sequence from genome browser and Ensemble (which include reference sequence of the exons) identify mutations that are responsible for CTLN1 disease in the families (Figure 3.5-3.8).

Figure 3.1 Pedigree of Saudi Arabian families segregating CTLN1 in an autosomal recessive fashion. A represent the first family, B represent the second family, circle represents female individuals, while square represents male individuals. Filled symbols represent affected members.

Figure 3.2 1% Agarose gel electrophoresis pattern of PCR amplified coding exons of the ASS1 gene for the first family, patient (III:3). Lane I (ladder size= 10bp) , lane 2-15 (exons 2-15), the size of exon 7 (300bp).
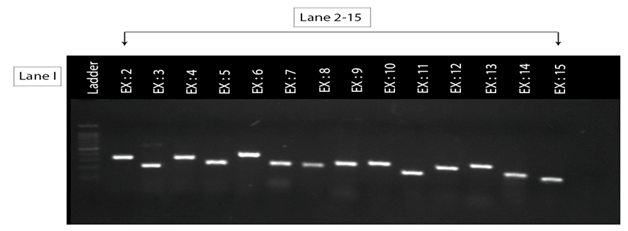
Figure 3.3 1% Agarose gel electrophoresis pattern of PCR amplified coding exons of the ASS1 gene for the second family patient (III:1). Lane I (ladder size= 10bp), lane 2-15 (exons 2-15), the size of exon 5(350bp).

Figure 3.4 1% Agarose gel electrophoresis pattern of PCR amplified coding exons of the ASS1 gene for unaffected individuals, Lane I (ladder size=10bp) lane 2 (exon 7) for the first family (II:2) lane 3 (exons 5) for the second family (II:2), the size of exon 5 and 7 (250bp).
Family one: Affected member 3 of third offspring of first family (III:3) (pedigree 3.1)
Exon (7-forward)
C>T


Figure 3.5 Electrochromatograms of one coding exon of ASS1 gene sequenced in citrullinemia patient (III:3). Arrow head indicate point of variant.
Carrier member 2 of second offspring of first family (II:2) (pedigree 3.1)
Exon (7-forward)
C>T


Figure 3.6 Electrochromatograms of one coding exon of ASS1 gene sequenced in citrullinemia patient (II:2). Arrow head indicate point of variant.
Family two: Affected member 1 of third offspring of second family (III:1) (pedigree 3.1)
Exon (5-forward)
A>G
Citrullinemia type 1 is an inherited autosomal recessive disease. It is a member of urea cycle disorders. It is characterized by accumulation of ammonia in the blood because of defect in enzyme ASS, which is either being missing or not working properly.2 The Prevalence of CTLN1 among the world has been estimated to occur in 1:57,000 births11 ASS1 gene is the responsible gene of producing argininosuccinate synthase 1 enzyme. Thus, the mutations in this gene lead to CTLN1.
In the present study, which is the first study on ASS1 gene carried out in Al-Madinah Al-Monawarah, two families were investigated with three generations autosomal recessive CTLN1 and severe phenotypic expression of the disorder. The 14 exons of the ASS1 gene -which is the gene that is responsible for CTLN1- was screened in two families. Missense and splice site mutations were found in affected individuals of family 1 and 2 respectively. In the affected individual (III:3) of first family, the homozygous variant was in exon 7 (c.501 C>T, p.166 His>His). The mother (II:2) of this affected individual was also found to be homozygous for same variant (c.501 C>T, p.166 His>His). While the affected individual (III:1) of second family has a splice site homozygous mutation affecting exon 5 (c.364-2 A>G). The mother has a heterozygous mutation in exon 5 (c.371 A>T).
ASS1 gene transcribe into a mRNA of 56,568 bp. It translates into a protein product of 412 amino acids (UCSC, 2017).25 Previous study conducted in China found missense mutation located in exon 13 (c.970 G>A, p.223 Gly>Ser).31 Another study conducted on Indian people found missense mutation in exon 3 (c.835 C>T, p.279 Arg>stop codon).32 ASS1 gene harbor different mutations and the affected individuals show variable phenotype based on lifestyle, race and population. Previous review of mutations in the ASS1 gene listed 87 mutations, including 27 novel mutations in patients with citrullinemia. Mutations are distributed throughout the gene, and it is usually difficult to predict the phenotype based on genotype. However, the p.G390R mutation in exon 15 was found to be the single most common mutation in patients with the classic phenotype 33 Another study found that since most patients with citrullinemia express stable mRNA in fibroblasts, the disorder is ideally suited for gene amplification with PCR and sequence analysis of mutant cDNA. They sequenced cDNA from 11 independent chromosomes and identified 9 different mutations: 3 showed absence of exon 5, 6 or 7, and 6 showed point mutations. Five of the 6 involved G>T transitions in CpG dinucleotides, and 3 of these resulted in loss of MspI sites.34
In the course of studying the molecular nature of mutations in Japanese patients with classic citrullinemia, researchers found that 10 of 23 affected alleles had the same mutation, i.e. deletion of exon 7.35 This differed from the situation in the United States, where far greater heterogeneity of mutations had been found. Another study reported that 20 mutations had been identified in ASS1 mRNA in classic citrullinemia, including 14 single base changes causing missense mutations, 4 mutations associated with an absence of exons 5, 6, 7, or 13 in mRNA, 1 mutation with a deletion of the first 7 bases in exon 16 (caused by abnormal splicing), and 1 mutation with an insertion of 37 bases between the exon 15 and 16 regions of mRNA.36–38 Moreover, a splice site mutation (IVS6-2A>G) has also been reported in 23 families (20 from Japan and 3 from Korea).
Mutations of The ASS1 Gene: (Table 3.1)
Location |
Protein |
mRNA |
DNA |
Ethnicity |
|
Nonsense |
|||||
Int 0 |
c.-4C4T |
Germany |
|||
Ex 7 |
p.G156X |
c.459_466del |
Germany |
||
p.N158X |
c.450_451delCT |
Switzerland |
|||
Int 11 |
r.773_774ins47 |
c.773149C4T |
Germany, Turkey |
||
Ex 12 |
p.G275X |
c.823G4T |
Netherlands |
||
Ex 12 |
p.R279X |
c.835C4T |
USA |
||
Ex 13 |
p.Q311X |
c.931C4T |
Italy |
||
Ex 14 |
p.R344X |
c.1030C4T |
Germany |
||
Ex 15 |
p.Q380X |
c.1138C4T |
Greece |
||
Int 15 |
p.Q401X |
c.1194-21_1213del |
Mexico |
||
Splice site |
|||||
Int 4 |
c.17411G4T |
Netherlands |
|||
Int 6 |
c.421-2A4G |
USA |
|||
Int 7 |
r.421_495del |
c.49511G4T |
Scotland |
||
Int 11 |
c.77311delG |
Japan |
|||
Int 11 |
c.77312_77313insT |
Spain |
|||
Int 12 |
c.83811G4A |
France |
|||
Int 13 |
r.839_970del |
c.97015G4A |
Japan |
||
Int 14/ Ex 15 |
r.1127_1128ins67 |
c.1127-9_1185dup67 |
Japan |
||
Int 15 |
r.Ex16del7 |
c.1194-1G4C Turkey |
Turkey |
||
Int 15 |
p.L399AfsX409 |
r.1193_1194ins37 |
Japan |
||
Deletion |
|||||
Ex 5 |
p.R100PfsX139 |
c.299delG |
Japan |
||
Int 7/Ex 8 |
c.496-21_515del |
Italy |
|||
Ex 11/Ex 12 |
r.689_838del |
Poland |
|||
Ex 13 |
p.E298RfsX315 |
c.892delG |
Spain |
||
Ex 13 |
p.A318LfsX375 |
c.952delG |
Germany |
||
Missense |
|||||
Ex 3 |
p.G14S |
c.40G4A |
USA |
||
p.S18L |
c.53C4T |
USA |
|||
p.C19R |
c.55T4C |
Japan |
|||
Ex 4 |
p.Q40L |
c.119A4T |
Germany |
||
Ex 5 |
p.V69A |
c.206T4C |
Spain, Germany |
||
p.S79P |
c.235T4C T |
Thailand |
|||
p.R86C |
c.256C4T |
USA, Italy |
|||
p.R86H |
c.257G4A |
Japan |
|||
p.R95S |
c.285G4T |
Turkey |
|||
p.P96S |
c.286C4T |
Turkey |
|||
p.P96H |
c.287C4A |
Italy |
|||
p.R108L |
c.323G4T |
Spain, Germany |
|||
p.G117S |
c.349G4A |
Austria, Thailand |
|||
p.G117D |
c.350G4A |
Spain |
|||
p.A118T |
c.352G4A |
Japan, Morocco |
|||
p.T119I |
c.356C4T |
Spain |
|||
Ex 6 |
p.D124N |
c.370G4A |
Netherlands |
||
p.R127W |
c.379C4T |
UK/Arabic |
|||
p.R127Q |
c.380G4A |
USA, Germany |
|||
Ex 7 |
p.R157C |
c.469C4T |
Japan |
||
p.R157H |
c.470G4A |
USA |
|||
p.L160P |
c.479T4C |
Germany |
|||
Ex 8 |
p.W179R |
c.535T4C |
Turkey, Italy, Libya. |
||
p.S180N |
c.539G4A |
USA |
|||
p.Y190D |
c.568T4G |
Germany |
|||
Ex 9 |
p.E191K |
c.571G4A |
Israe |
||
p.E191Q |
c.571G4C |
N. Ireland |
|||
p.A192V |
c.575C4T |
Japan |
|||
Ex 10 |
p.A202E |
c.605C4A |
Germany |
||
p.L206P |
c.617T4C |
Germany |
|||
Ex 12 |
p.V263M |
c.787G4A |
Turkey |
||
p.R265C |
c.793C4T |
India |
|||
p.R265H |
c.794G4A |
Japan, Germany |
|||
p.V269M |
c.805G4A |
Germany |
|||
p.E270Q |
c.808G4C |
Spain, Germany |
|||
p.R272C |
c.814C4T |
Japan |
|||
p.K277T |
c.830A4C |
France |
|||
p.R279Q |
c.836G4A |
Japan |
|||
p.G280R |
c.838G4C |
Japan |
|||
Ex 13 |
p.E283K |
c.847G4A |
Belgium |
||
p.T284I |
c.851C4T |
Germany |
|||
p.Y291S |
c.872A4C |
USA, Germany |
|||
p.D296G |
c.887A4G |
Italy |
|||
p.M302V |
c.904A4G |
Italy |
|||
p.R304W |
c.910C4T |
Japan, Turkey |
|||
p.R307C |
c.919C4T |
Germany |
|||
p.K310Q |
c.928A4C |
Canada |
|||
p.K310R |
c.929A4G |
USA |
|||
p.G324S |
c.970G4A |
USA, Korea, Turkey |
|||
Ex 14 |
p.G324V |
c.971G4T |
Mexico |
||
p.S341F |
c.1022C4T |
Turkey |
|||
p.V345G |
c.1034T4G |
Germany |
|||
p.G347R |
c.1039G4C |
Spain |
|||
p.Y359D |
c.1075T4G |
Germany |
|||
p.G362V |
c.1085G4T |
Netherlands, Turkey, Germany |
|||
p.R363W |
c.1087C4T |
USA, Germany |
|||
p.R363G |
c.1087C4G |
Greece |
|||
p.R363L |
c.1088G4T |
Japan |
|||
p.R363Q |
c.1088G4A |
USA |
|||
Ex 15 |
p.T389I |
c.1166C4T |
Canada |
||
p.G390R |
c.1168G4A |
USA, Spain, Turkey, Bolivia, |
|||
Table 3.1 Mutations of the ASS1 Gene
From the present study, the following could be concluded: Mutations in ASS1 gene were found using the sequences analysis. In the first family affected member III:3 and unaffected member II:2 have a variant in exon 7 (c.501 C>T, p.166 His>His).
In the second family affected member III:1 has a splice site mutation in Exon5 (c.364-2 A>G). The splice site region is important because mutations in these areas may lead to entire exon being spliced out of the mRNA. The unaffected member II:2 of the second family has a heterozygous mutation in exon 5 (c.371 A>T). A heterozygous mutation makes individual a carrier of the disease.
Ethical approval: The research have two ethical approvals. The first from Taibah University. And the second from Ministry of Health
First of all, praise is due to almighty ALLAH with his compassion and mercifulness to allow us completes this work.
Thanks and sincere respect to Dr. Samia Mahdi Ahmed Mohammed for her guidance and supervision this study.
Our special thanks and respect for Dr. Waleed Mahlawi (Dean of Applied Medical Sciences College) and Dr. Walaa Mohammed Ahmed for their valuable help and support, and enabled us to complete this research.
Our thanks for Dr. Mohammed Samman (Directer of Center of Genetics and Inherited diseases), Dr. Sulman Basit and Alia Albalawi for helping us to carry out this research at the center.
Our thanks also for Dr. Zakaria Hawsawi, Dr. Om-hani Malibari, Yasir Al-Mawled, Heba Al-shenqiti, and Salma Alsayed for their collaboration and support.
Finally, we would like to express our sincere thanks and gratitude to the patients and their families for their collaboration.
Words can never express our feelings towards everyone stood beside us in this research, from the initial stages until the final draft.
Author declares there are no conflict of interests.

©2018 Mohammed, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.