International Journal of
eISSN: 2573-2889


Research Article Volume 6 Issue 1
Department of Biotechnology, Akwa Ibom State University, Uyo, Nigeria
Correspondence: Elijah Nya, Department of Biotechnology, Akwa Ibom State University, Nigeria, Tel +234 8125344294
Received: August 24, 2023 | Published: August 31, 2023
Citation: Elijah N, Atim P, Ofonime U. Profiling of fungi associated with Lycopersicum esculentum mill. spoilage in Nigeria using internal transcribed spacer (its) sequence of rDNA method. Int J Mol Biol Open Access. 2023;6(1):25-30. DOI: 10.15406/ijmboa.2023.06.00149
Lycopersicum esculentum Mill often referred to as Tomato is among important vegetable fruits widely cultivated the world over. The fruits are made up of soft endocarp which make them susceptible to spoilage by fungi mold. The fungi produce mycotoxins that are injurious and detrimental to human health. This study is aimed at profiling fungi mold associated with Lycopersicum esculentum rot /spoilage and identify the putative fungi responsible for its spoilage in Nigeria, particularly Akwa Ibom State metropolis. Tomato fruits heavily infested with fungi mold were collected and marshed using a sterile electric blender. Serial dilution of sample in normal saline were made and grown on prepared plates of Potato Dextrose Agar (PDA) to obtain pure culture. High quality genomic DNA was isolated from the samples; it was quantified and checked for purity. The DNA samples were subjected to PCR amplification. The amplicons were gel eluted, sequenced and checked for homology by using Basic local alignment search tool - BLAST. Identification was obtained from the top similarility outcomes of the samples. Four fungal isolates identified were Aspergilus spp., Fusarium spp., Mucor spp. and Penicillium citrinium. The economic importance of fungi molds are not in doubt as some of them are important precursor of diseases as they produces mycotoxins which is capable of causing diseases in both humans and animals, while other sare important in the field of medicine, in industrial production of antibiotics especially Penicillin.
Keywords: profiling, lycopersicum esculentum, internal transcribed spacer sequence of rDNA, blast
Lycopersicon esculentum Mill is important vegetable fruit cultivated and consumed the world over. Tomato is an annual plant belonging to the Solanaceae family, others vegetables in the same plant family includes Pepper, Potato, Tobacco and Spinach; Egg plants Solanum melongena or Aubergine and can grow to the height of over one to two meters. The fruit is a berry, grown mainly in arable lands, gardens, with little or no irrigation in Nigerian soil.1
Lycopersicon esculentum fruit is rich in vitamins, essential minerals and lycopene, which is excellent antioxidant and so play a vital role in human nutrition.2–4 As an important antioxidant, it serves to lessen the risk of prostate and breast cancer5 and keep a good body and normal healthiness. The cultivation of this vegetable is highly cherished that about one million hectares of arable land is devoted for it cultivation every year.6,7 Nigeria is the second largest producer of this vegetable fruit in Africa,7 and it accounts for about 18% of the average daily intake in Nigeria.8 Tomato may be used in the production of fruit drinks or processed into tomato - pastes which are used for thickening of stews.9
Interestingly, the limiting factors with this tomato berry, which of course, negatively affect the fruit economic value, is the inadequate storage facilities and relative inherent short shelf-life caused by ubiquitous pathogenic microorganisms in the environment, causing myriad of attacks and thus leading to spoilage with attendant deterioration in the quality of the tomato fruits.10 Even in the northern region of the country where most tomato vegetables are grown, there are no records of any postharvest storage facilities available or organized traditional storage methods for tomato fruit.11
Furthermore, to stem the tides of this drawback, they are usually sold immediately after harvesting, packed in baskets, cardboard boxes, or wooden crates and transported to the markets. The fruit is the most perishable portion of the vegetable. This is because tomato has high moisture contents which are also responsible for their being susceptible to spoilage organisms such as bacteria, fungi and protozoans.12 Most authors have reported estimates of losses due to these vagaries; about one third of the produce is lost before reaching the end users. Principally, this loss has also been implicated in number of factors including mechanical damage and bruises inflicted by pathological agents, thus reducing the market value of the tomato to barely minimum.
Garcha13 regards spoiled food as food items that has been injured and cause to be unfit for human consumption. Other factors have been implicated in food spoilage, a part from microbial activities causing enzymatic changes; they include insect activities causing physical bruises and changes in taste, texture, colour, smell and quality deterioration. This spoilage of fruits is not deliberate but it is a normal ecological function of these elements. It has also been estimated by Garcha13 that 20% of all fruits and vegetables harvested for human consumption are lost through microbial spoilage. Spoilage of fresh tomato usually occurs during transit and storage as they, after picking continue to respire producing unpredictable degrees of vital heat. Vegetables or fruits that generate the majority vital heat are those that get rotten most readily.14
However, Penicillium, Mucor, Fusarium and Aspergillus are common fungi that cause similar wilt disease of tomato.15 Phytophthora capsici – as soil borne fungi is well documented16 Fusarium is more common in replanted fields and gardens, while Verticillium has been very destructive on greenhouse and in commercially stake tomatoes. Verticillium wilt is more of a problem at temperate regions while Fusarium wilt is considered a tropical disease where temperature is warmer. Most fungi are common soil borne.17 These soil borne fungi causes severe losses in tomato, they include Alternaria solani known to be responsible for early blight disease of tomato, Septoria lycopersici causes Septoria leaf spot of tomato, Fusarium oxysporum causes Fusarium wilt.These fungi become established mostly in moist soils and can remain in the soil for years. When tomatoes are planted in infected soil, they become attack through the roots. This present study lend importance on the spoilage fungi of Lycopersicon esculentum. Profiling and identification of the pathogenic fungi species associated with tomato will help in many ways to proffer and decipher their control strategies in Nigeria.
Fresh ripened tomatoes were purchased from Ikot Akpaden Junction and were kept for one week to spoil and overgrown with mold mucilage. The spoilt tomatoes were then transported to microbiology laboratory in Akwa Ibom State University for analysis. The tomatoes were marshed using a sterile electric blender and 1 gram of the sample was weighed and suspended in 9ml of normal saline for serial dilution assay.
Isolation of fungi
A serial dilution of 101–5 was carried out for the samples. After the dilution, a drop from dilution factor 103, 104 and 105 were plated out into petri-dishes of appropriate Agar medium (potato dextrose agar). The agar plates were then incubated at room temperature for 2-5 days and observed for growth.
Purification of fungal isolates
During the seventy-two (72) hours of incubation, plates were observed for obvious growth and plate that displayed growth were marked and sub-culture to obtain pure colonies. After obtaining the pure colonies, they were preserved in sterile Mc Cartney’s bottle for further use. The stock bottle were kept in a refrigerator at 40C
Identification of isolated fungi
Profiling for identification of the pure Isolates was done morphologically at the initial time. Morphological characterization was done using a 7 day pure fungi cultures cultivated at 25± 20C on PDA growth medium. Further to this, cultural and microphotographs were taken for future references and comparison for fungus identification. Macroscopic characters including colony color, size and shape were studied ocularly intadem with the microscopic characters such as conidiophores, length, wall, vesicle size, and shape which were also examined under microscope.
For microscopic examination of each fungal isolate, small pieces of fungal colony were smeared on a slide and focus on 40 magnification lense of a calibrated compound microscope. Complete description of each isolate based on macro and micro morphological characters observed was arranged. Fungal isolates were splited out by its descriptive comparison according to Thom18 Their common genotypic and phenotypic characters were confirmed molecularly by using Internal Transcribed Spacer ITS sequence analysis of ribosomal DNA rDNA
Extraction of total DNA
Fungal Genomic DNA was extracted with C-TAB Cetyl trimethyl ammonium bromide, however, with some modifications. Briefly, 1,200μl of recent pure fungal cultures were collected in 1.5ml Zymo Research ZR Bashing Bead™ Lysis Eppendorf tubes containing Zirconia beads and stored in a -80°C freezer (U410 New Brunswick high efficiency freezer) to freezed dry overnight. The ZR Bashing Bead™ Lysis Tube was centrifuged in micro centrifuge at ≥10,000g for 1 minute. Upto 400μl supernatant was transferred to Zymo-Spin™ IV Spin Filter (orange top) in a collection tube and centrifuged at 7,000 rpm (~7,000g) for 1 minute. The base of the Zymo-Spin IV™ Spin Filter was snapped off prior to use. 1,200μl of fungal rDNA, binding buffer was added to the filtrates in the collection Tube. The beads were removed with the aid of a magnet and 500µl of CTAB extraction buffer was added to the tube and vortex for a moment. The mixture was then incubated on an Eppendorf thermomixer F1.5 at 65°C for 1h. After incubation the mixture was centrifuged for 10 mins at 15,000 rpm. The upper phase was carefully extracted and transferred to a fresh tube, then 500µl of chloroform: isoamyl (24:1) was added and mixed by inverting the tubes. The mixture was centrifuged at 15,000 rpm for 10mins and the upper phase extracted and transferred to fresh tubes. This step was repeated and at last, 350µl of 2-propanol was added, mixed again by inversion and the mixture was centrifuged at 15000 rpm for 10mins. After centrifugation a clear white pellet appeared at the bottom of the tube, the aqueous phase was carefully decanted to avoid losing the pellet. Approximately 500µl of 70% ethanol was added to the tubes and gently tapped with fingers to release the pellet, then centrifuged for 10 mins at 15000 rpm. The aqueous phase was decanted carefully and the pellet allowed to dry for 30mins in open air. Approximately 30µl of rDNA Elution Buffer (Tris-EDTA) TE buffer was added to the rDNA directly and Ultra-pure rDNA was then obtained and stored in a Liebherr medline -20°C freezer until required for use during the next experiment.
PCR amplification
The amplification experiments were carried out using a cooling and heating thermal cycler. The amplification reactions were conducted using 0.5µl forward and 0.5µl reverse primers of the ITS gene. ITS (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS 4 (5’-TCC TCC GCT TAT TGA TAT GC-3’) were carried out using the following condition: 30 cycles of denaturation at 94oC for 30 seconds, an initial elongation at 68oC for 1 minute and elongation at 68oC for 5 minutes.
Agarose gel electrophoresis of PCR products
The Agarose gel electrophoresis of PCR products was carried out according to Nya19, however with slide modifications. Briefly, the Amplicons were separated on 1.5% Agarose gel. The Agarose gel of 1.5% was prepared by dissolving 1.5 g of Agarose in 100ml of Tris-EDTA (TE) buffer and heated in a microwave oven for 4 to 5mins. The mixture was allowed to cool briefly and then 6µl of ethidium bromide stain was carefully added to the mixture and mixed by gentle shaking in a fume hood. The mixture was poured into a gel casting tray mounted with combs and allowed to solidify. The gel was then transferred to a horizontal gel electrophoresis tank containing 1X TE buffer and the combs gently removed. Approximately 1.5µl of loading dye was added to the PCR products. The mixture was briefly vortexed and gently loaded onto the gel inside the tank. Six 6 µl of rDNA ladder of 100bp was used as molecular weight standard. This was also loaded on the first well and the set-up was allowed to run at 130V for 20 minutes.
Electrophoresis was viewed on the UV trans-illuminator which showed about 500bp PCR products.
PCR products purification for sequencing
The PCR products were purified in four main steps. The first step was the adjustment of the bonding conditions whereby 110µl of bonding buffer B3 was added to 22µl of the PCR product and the sample was mixed. The mixture was then transferred to EZ-10 spin column (PCR product purification Kits), incubated for two minutes, and there after centrifuged at 9000rpm for 30 seconds and the flow was discarded. Wash solution (500µl) was added to the spin column and centrifuged at 9000rpm for 30 seconds and the flow discarded. The process was repeated for 1 minute. Elution of the genomic rDNA was the last step where the collection tube was replaced with a clean 1.5µl of 65oC elution buffer was added and the mixture incubated at room temperature for 2 minutes. The mixture was centrifuged at 9000 rpm for 1 minute followed by removal of the EZ-10 spin column. The sample was stored -20oC for further use.
Photography and documentation
This was done according to a method used by Nya19 Briefly, after electrophoresis the gel was transferred to a High performance UV transilluminator (UVP, United Kingdom), and viewed with a safety google and the pictures taken with EOS 1200D camera (Canon, Taiwan).
rDNA Sequencing and bio-informatics
The stored purified rDNA products were sequenced with the same primers used for PCR amplification. Sequencing was done using rDNA Sequencer: IIIumina Genome Analyzer IIX, and the resulting raw data was edited with Chromas rDNA sequencing software for generation of a consensus sequence. The consensus sequence in FASTA format were used for identification of the microorganism using Basic Local Alignment Search Tool BLAST, to compare the consensus sequence with published sequences in the Gene bank of National centre for Biotechnology Institute NCBI. Distribution of top 100 BLAST hits on 100 subject sequence
Morphological description of the four (4) fungal isolates (Aspergilus sp, Fusarium sp, Mucor sp and Penicillium sp)
The four (4) fungal isolates that were isolated from heavily fungi infested tomato collected within Akwa Ibom state were examined for morphological variations. A 7 day old pure fungus cultures grown at 25 ± 20C on PDA growth medium were macroscopically and microscopically examined, photographed and documented as seen in Figure 1: a,b,c and d. Morphologically, most of the fungi showed characters like colony color, size, shape as observed visually and microscopic characters manifested to include distinct variations with respect to the conidiophores, length, wall, vesicle size, and shape
Molecular study of rDNA.
Ribosomal DNA (rDNA) extraction was carefully undertaken following the extraction procedures earlier stated, the rDNA was observed to be present as a colorless pellet at the bottom of the Eppendorf tubes.
rDNA Quality analysis on gel electrophoresis
The rDNA extracts from the samples were analyzed in a 1.5% Agarose gel with 1x of Tris-EDTA (TE) buffer, for bands purity estimation under UV- light and documented using the gel documentation station (Aplegen- Eduro GDS Gel documentation Beckman system). DNA ladder of 500bp (New England Biolabs Inc., UK), DNA marker (100-1000bp) was used, the rDNA band was marked at 500bp on the DNA ladder.
DNA Sequence of the amplified product and bioinformatics
The amplicons obtained from the Internal transcribed spacer ITS was further sequenced using 0.5ul forward and 0.5ul reverse primers of the ITS gene. The nucleotide sequence obtained was not aligned at both ends. DNA sequence length of 500bp for the rDNA forward sequence and 100bp for reverse sequence length respectively. The sequence information was uploaded into the GenBank database and the identification was obtained from the top similar outcomes as shown in Figure 1a,b,c and d. The blastn sequence search for forward and reverse sequence of fungi samples, indicating the outcome of similarities with their % identities are shown in Figure 3 & Table 2.
Profiling of four (4) fungal isolates (Aspergilus sp, , Mucor sp, Fusarium sp and Penicillium sp) were carryout from the heavily infected spoilt tomato samples. Distinct characters among the four (4) fungal isolates were investigated using morphological descriptors in line with Nya and Eka.20 The four isolates were examine for morphological distinction based on the expressed phenotypic attributes of the fungi isolates. The fungal samples obtained from different heavily infested tomato were different from each other from the perspective of their morphological appearance, size and colour as shown in Table 1 and reinforced in the results of phylogenetic tree (Figure 6) demonstrating the evolutionary relationships of the taxa. The most frequently encountered fungi of tomato contamination i.e. Penicillium spp. (Penicillium citrinum, Talaromyces ruber), Aspergillus spp, Mucor spp, and Fusarium spp, could probably be due in part, to the fact that Tomato Fruits, by nature have low pH, higher moisture content and these inherent attributes renders them very susceptible to attack by disease pathogens, especially fungi, which in addition to causing rots may also produce mycotoxin, thus making them unfit for human utilization.21–23 Secondly, this work showed alignment with Adeoye24 who also affirmed fungal prevalence in moist environment and attack on tomato is alarming, Sommer, (1985) reported Mucor spp, and Fusarium spp. as causing a wide range of fruits disease resulting in destructive and economic allosses during marketing, transportation and storage or preservation.
|
Isolates Identity |
Texture |
Hyphae |
Special Vegetative Structure |
Surface Colour |
Shape |
Organism |
|
Isolate a |
Velvety |
Septate |
Foot cell |
White and typical black spores |
Globose |
Aspergilus spp. |
|
Isolate b |
Cottony |
Septate |
Curved |
White wooly |
Sickle- shape |
Fusarium spp. |
|
Isolate c |
Cottony
|
Non-Septate |
Mycelium |
Pale grey to yellowish brown |
Globose |
Mucor spp. |
|
Isolate d |
Velvety |
Septate |
Multicellular mycelium |
Pale yellow to reddish-brown |
Globose |
Penicillium sp.
|
Table 1 Morphological and Cultural Characterization of fungi isolates obtained from the infected spoilt Tomato samples
|
Score |
Expect |
Identities |
Gaps |
Strand |
|
780 bits(864) |
0 |
446/446(100%) |
0/446(0%) |
Plus/Minus |
|
Query 1 |
GGGGGTCGGCTGGCGCCGGCCGGGCCTACTAGAGCGGGTGACGAAGCCCCATACGCTCGA |
60 |
||
|
Sbjct 480 |
GGGGGTCGGCTGGCGCCGGCCGGGCCTACTAGAGCGGGTGACGAAGCCCCATACGCTCGA |
421 |
||
|
Query 61 |
GGACCGGACGCGGTGCCGCCGCTGCCTTTCGGGCCCGTCCCCCCGGCgggggggACGGGG |
120 |
||
|
Sbjct 420 |
GGACCGGACGCGGTGCCGCCGCTGCCTTTCGGGCCCGTCCCCCCGGCGGGGGGGACGGGG |
361 |
||
|
Query 121 |
CCCAACACACAAGCCGGGCTTGAGGGCAGCAATGACGCTCGGACAGGCATGCCCTCCGGA |
180 |
||
|
Sbjct 360 |
CCCAACACACAAGCCGGGCTTGAGGGCAGCAATGACGCTCGGACAGGCATGCCCTCCGGA |
301 |
||
|
Query 181 |
ATACCAGAGGGCGCAATGTGCGTTCAAAGACTCGATGATTCACTGAATTCTGCAATTCAC |
240 |
||
|
Sbjct 300 |
ATACCAGAGGGCGCAATGTGCGTTCAAAGACTCGATGATTCACTGAATTCTGCAATTCAC |
241 |
||
|
Query 241 |
ATTAGTTATCGCATTTCGCTGCGTTCTTCATCGATGCCGGAACCAAGAGATCCGTTGTTG |
300 |
||
|
Sbjct 240 |
ATTAGTTATCGCATTTCGCTGCGTTCTTCATCGATGCCGGAACCAAGAGATCCGTTGTTG |
181 |
||
|
Query 301 |
AAAGTTTTAAACTAATTTCGTTATAGGTCTCAGACTGCAACTTCAGACAGCGTTCAGGGG |
360 |
||
|
Sbjct 180 |
AAAGTTTTAAACTAATTTCGTTATAGGTCTCAGACTGCAACTTCAGACAGCGTTCAGGGG |
121 |
||
|
Query 361 |
GGCCGTCGGCGGGCGCGGGGCCCGCCGAGGCAACATAGGTTCGGGCAACACGGGTGGGAG |
420 |
||
|
Sbjct 120 |
GGCCGTCGGCGGGCGCGGGGCCCGCCGAGGCAACATAGGTTCGGGCAACACGGGTGGGAG |
61 |
||
|
Query 421 |
GTTGGGCCCCGAGGGGCCCGCACTCG |
446 |
||
|
Sbjct 60 |
GTTGGGCCCCGAGGGGCCCGCACTCG |
|
35 |
|
Table 2 Results for alignment of query sequence with the subject in the gen bank (NCBI)
Furthermore, the findings from this study showed that tomato fruit are susceptible to several fungi since fungal contaminations of tomato are common in tropical countries. This is in line with many authors who have attributed this to improper handling, mechanical injuries such that occur during harvesting, which could be window for an infected sites for spoilage fungi. They also implicated post harvest storage aggravated by lack of good storage facilities, the ability of the fungi to produce spores and their ubiquitous nature, couple with environmental conditions such as temperature and relative humidity. These all contribute to the severity of this problem in the tropical
environment.25–27
However, after morphological identification of the fungi isolates using a microscope, each specie like Penicillum citrinum (Figure 1) was taken for molecular profiling or characterization. Molecular profiling provide valuable information on genetic diversity putative organisms in view of its ability to decipher variation among organisms existing at the rDNA level. Profiling of any organisms up to species level is of fundamental importance for rapid identification and diversity studies.28 This showed that for the profiling of the fungal diversity, it is essential that individual fungus be classified accurately to species level. Furthermore, to establish effective management of tomato fruit against diversities of fungi attack, it is imperative to go as far as molecular profiling, with which genetic variations could be detected. The genetic compositions of organism are always distinct. In view of this, their discrimination from among numerous types and their wild relatives becomes necessary.
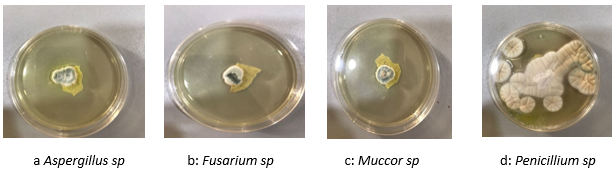
Figure 1 morphological variation among the 4 fungal isolates of tomato as observed with the naked eye.
Principally, the genetic composition of the fungi species studied have been shown to be different, indicating that the perceived observable differences phenotype are true and are not responsible or due to environments but rather to genetic component Genotype. As a result, their identification from among others has become essential especially in germplasm study and for the formulation of appropriate post harvest management plan.22,24 Furthermore, rising from our perceived focus and the objectives of our study, molecular profiling of fungi associated with tomato spoilage, this may be considered as fundamental important for generating new tomato variety with improved post harvest quality for tackling global food insecurity.
rDNA obtained from the fungal samples were analyzed on Agarose gel electrophoresis for purity assessment under UV- light and before going for the gel documentation station for record documentation using Aplegen- Eduro GDS Gel documentation Beckman system. However, 50bp rDNA ladder New England BiolabsInc., UK, DNA marker (50 - 1000bp) was used and the rDNA band was marked at 500bp on the rDNA ladder. Amplification using the pairs of primers ITS1 and ITS4 (Figure 2) was carried out and UV Trans-Illuminator showed PCR product at about 500bp and the raw rDNA higher up the gel due to differences in molecular weight, M (Gel ladder), B (PCR product) and RD (Raw rDNA) (Figure 2). The results display that the fungi isolates were effectively amplified and was shown as clear bands at 500bp, this was inspite of the differences in the location where the samples were obtained and distances from each other. Thus, any variations could be due to genetic constitution of individual strain. Interestingly, this findings necessitate the calls for further study to investigate the genetic consistency of the different isolates. Chromatogram peaks stained with fluorescent dye showed the arrangement of nucleotides in the rDNA sequence and each colour represented a specific nucleotide (Figure 3).
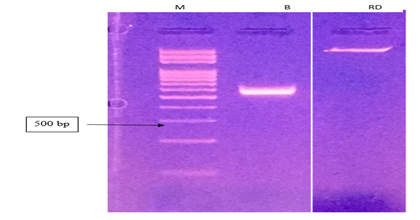
Figure 2 DNA gel image of the fungi associated with tomato as shown on the gel documentation (Aplegen- Eduro GDS Gel documentation Beckman system)
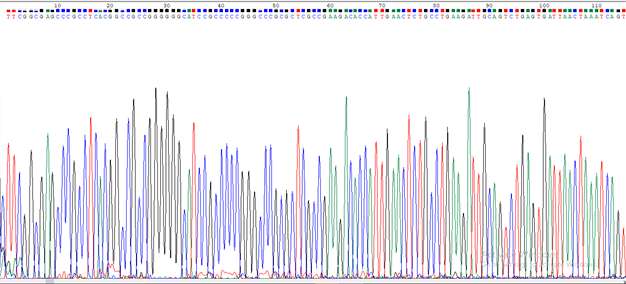
Figure 3 chromatogram peaks stained with fluorescent dye showing the arrangement of nucleotides in the DNA sequence
BLAST performance: For the sequences similarity alignment checks with those in the NCBI database, numerous species were obtained as being similar. This is as shown in Figure 4. This shows that some of the species were similar with those of the AB-forward sequences obtained from the NBCI The BLAST revealed ITS sequence of all strains showing 97−100% identity similarity with those strains stored at GenBAnk (Figure 4). Distribution of the first 100 strains in the basic local alignment search tool BLAST punch at 100 subject sequences and the red lines signified an alignment score between the query sequence and subject >200. The outcome of the Sequencing and basic local alignment search tool BLAST analysis of spoilt fungi showed dissimilarity among the different isolates. Invariably, showing that the fungi associated with tomato spoilage were phenotypically and genotypically disparate and that any morphological differences observed, were customarily due to their inherent genotypic constitutions without any environmental attributes. The non- similarity in samples genotypic components reported here among the fungi associated with tomato spoilage is in line with those reported earlier by Mehrotra and Ashok29 in other fruit vegetables predominantly in tomatoes, pepper, Spinach; Egg plants Aubergine or Solanum melongena.29,30 The account of rDNA ITS loci as examine and shown by PCR result and that of rDNA sequencing for the disparity checks and validation of fungi associated with tomato spoilage has established in an unmistakable term the discrimination and similarity of these all important fungi.The history of the samples and their evolutionary trends were subjected upon the UPGMA method.31 The phylogenetic tree showing the sum of branch length = 0.54041570 is as indicated in (Figure 5) = Phylogenetic Tree. Evolutionary distance was calculated with the p-distance method according to Nei and Kumar,32 The analysis involved 10 nucleotides sequence; codon positions were 1st+2nd+3rd+Noncoding. All positional introns containing gaps and missing data were removed. Sum total of 433 positions were observed in the final dataset. Analyses according to Kumar33 were use mainly MEGA7. The results showed differences in the evolutionary relationships of the taxa indicating, however, that the rDNA and ITS outcome of the various sample isolates investigated showed variations noticed among the isolated strains indicating non compliance with environmental and geographical differences.
None.
The authors declared that there are no conflicts of interest.

©2023 Elijah, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.