International Journal of
eISSN: 2573-2889


Research Article Volume 1 Issue 1
1Department of Biotechnology, SRM University, India
2Graduate Institute of Environmental Engineering, National Central University, Taiwan
Correspondence: Munusamy Thirumavalavan, Department of Biotechnology, School of Bioengineering, SRM University, Kattankulathur-603203, India
Received: October 27, 2016 | Published: November 16, 2016
Citation: Prabaharan C, Thirumavalavan M, Pachaiappan R. Production of antioxidant peptides from Ferula asafoetida root protein. Int J Mol Biol Open Access. 2016;1(1):19-24. DOI: 10.15406/ijmboa.2016.01.00003
The Ferula asafetida (F. asafoetida) root exudates proteins were hydrolyzed using gastric digestive enzyme to identify the antioxidant peptides. F. asafoetida root protein was hydrolyzed using gastrointestinal enzymes (pepsin, Papain, trypsin and α-chymotrypsin) and purified by gel filtration chromatography(fast protein liquid chromatography, FPLC). The fractions F1, F2, F3, F6, F7, F8, and F9 were collected and screened for radical scavenging activity. Antioxidant activities of hydrolysate were evaluated using 2,2’-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), ferric reducing antioxidant power assay (FRAP) and superoxide radical scavenging activity in enzyme-linked immunosorbent assay (ELISA) multimode reader. Results showed that the activity of quenched free radicals (ABTS+, FRAP and superoxide) was altered in a concentration-dependent manner. Fraction F1 showed highest scavenging activity than that of other fractions and it showed comparatively enhanced reducing power activity with butylated hydroxyanisole. Among three assays, ABTS showed better result than that of others. Further studies are still needed to find out the amino acid sequence responsible for antioxidant activity.
Keywords: ferula asafoetida, enzyme hydrolysis, antioxidant peptides, FPLC, gastrointestinal enzymes, liquid chromatography, electrophoresis device, medicinal properties, digestive enzymes
In recent years, there has been much importance placed on finding novel therapeutic agents from F. asafoetidaplant and it’s common name is Asafoetida. It is commonly used as a traditional medicine in Iran, Afghanistan and India. It is herbaceous plant of the Umbelliferae family and its root contains oleo gum resin which has many medicinal properties. The resin is solid or semisolid with alliaceous odor and a bitter acrid taste, which comprises ferulic acid, umbelliferone, asaresinotannols, umbelliferone ethers, gums and volatile oils. The sulfides of volatile oils are responsible for the characteristic flavor of Asafoetida1 Asafoetida is commonly used for cooking purpose as it is an indigenous medicine which cures sporadic disorder and stimulates digestion. Figure 1 shows the schematic representation of plant Ferula asafetida and its various applications.The free radicals are the main cause for oxidative damage to biomolecule such as DNA, proteins and lipids, which leads to many chronic diseases such as atherosclerosis, diabetes, aging, degenerative diseases and mainly cancer.2-5 Formation of free radicals such as superoxide anion (O2-) radical and hydroxyl (OH) radical is an unavoidable consequence in aerobic organisms during respiration. Uncontrolled generated free radicals are very unstable, and react rapidly with the other groups or substances in the body, leading to cell or tissue injury. Numerous physiological and biochemical processes in the human body may produce oxygen-centered free radicals and other reactive oxygen species as byproducts.
In addition, numerous studies have revealed that uncontrolled lipid peroxidation is involved in the occurrence of numerous chronic diseases.6-8 There is still considerable argument regarding the direct intake of natural antioxidants has been associated with reduced risks of cancer, cardiovascular disease, diabetes, and other diseases related with ageing.9-13 Therefore, studying of natural plants specially bred for higher levels of antioxidant compounds is a realistic approach to increase dietary antioxidant intake. However such screening methods should be simple, inexpensive, rapidly performed, and provide a high degree of precision. Many studies have been investigated the antioxidant property of hydrolysates from different plant and animal sources like sunflower protein, alfalfa leaf protein, corn gluten meal, egg-yolk protein, algae protein, mushroom 14. The defensive effects of natural antioxidants in plants and animals are related to different constituents. These techniques have shown different results and no correlations were obtained and discussed in details. Hence, in this study we made an attempt to obtain such hydrolysates fromF. asafoetida root protein and to study their antioxidant activity using different methods. The biological activity of a peptide is widely recognized to be based on amino acid composition.15 The bioactive peptides are commonly made up of 3-20 amino acids per peptide, which are present in the large protein in inactive sequences and become active when they get hydrolyzed by digestive enzymes, microbial enzymes or during food processing.16 Enzymatic hydrolysis of proteins is one of the significant approaches used to discharge bioactive peptides and is widely applied to study functional and nutritional properties of protein sources.6 These peptides can be directly used in the formulation of functional foods and nutraceuticals to prevent damage related to oxidative stress in human disease conditions. The main objectives of the current study were to obtain a protein from the root exudates of the F. asafetida, enzymatic hydrolysis of such isolated protein using gastrointestinal enzymes and finally to investigate the antioxidant property of obtained peptides fromF. asafoetida root protein. This approach is simple, effective and inexpensive and in this work the antioxidant activity was directly correlated with concentration factor. Therefore, it would be an appropriate technique for determining antioxidant in F. asafoetida root extract.
Chemicals
Enzymes were obtained commercially as follows; Papain from papaya latex (Merck, activity: >30,000 USP units/mg of protein calculated using hemoglobin as a substrate), pepsin from porcine gastric mucosa (Calbiochem, activity: 3000 U/mg of protein calculated using hemoglobin as a substrate), trypsin from porcine pancreas (Sigma-Aldrich, activity > 10000 UN/mg of protein calculated using BAEE as a substrate) and α-chymotrypsin from bovine pancreas (Sigma-Aldrich, activity > 2000 - 3000 UN/mg of protein calculated using ATEE as a substrate). Superdex-G-30 matrix was obtained from GE Healthcare Bio-Sciences AB, Sweden. The reagents used for the antioxidant assays such as 2,2’-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), ferrozine, ferrous chloride, ferric chloride, (2, 4, 6- tripyridyl-s-triazine) (TPTZ) and butylated hydroxyanisole (BHA) were purchased from Sigma-Aldrich, USA. All other chemicals and reagents used were of the highest grade available commercially.
Plant collection and protein extraction
The root exudate of F. asafoetida was obtained from Iran market. Exudate (250 g) was soaked in distilled water overnight and stirred to obtain a consistent solution. Preliminary level of filteration was done to remove the larger particles and debris from the solution. The obtained filterate was centrifuged at 18000 rpm for 30 min, supernatant was collected and stored at 4°C. Precipitation of proteins was done using ammonium sulphate at 80% saturation and kept for precipitating overnight at 4°C. The precipitated proteins were separated through centrifugation at 10000 rpm for 30 min. The pellet was dissolved in 15 ml of water and subjected to dialysis using 3.5 kDa MWCO dialysis centrifugation tube (Merck Millipore, Germany). The precipitated proteins were lyophilised (Lyodel Freeze Dryer Model: DPRG-1GH) and stored at -20°C for further use. Protein concentration was estimated by the Bradford method using bovine serum albumin (BSA) as a standard protein. The absorbance of the protein-dye complex was measured at a wavelength of 595 nm.
Glycine SDS-PAGE
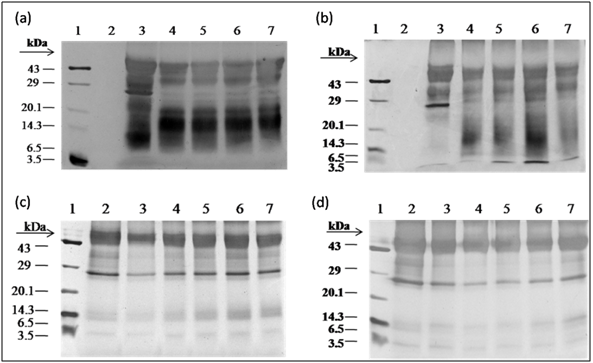
F. asafoetidaproteinswas separated through the glycine SDS-PAGE (14% separating gel and 4% stacking gel) in the electrophoresis device (GeNei, India). The separation was controlled with 100 V and allowed to run until the dye reaches the glycine SDS-PAGE bottom. The protein bands were stained with silver staining. Protein patterns were observed and documented using Bio-Rad (ChemiDoc XRS+ imaging system, USA) and compared with the protein marker 43 - 3.5 kDa.
Enzymatic hydrolysis
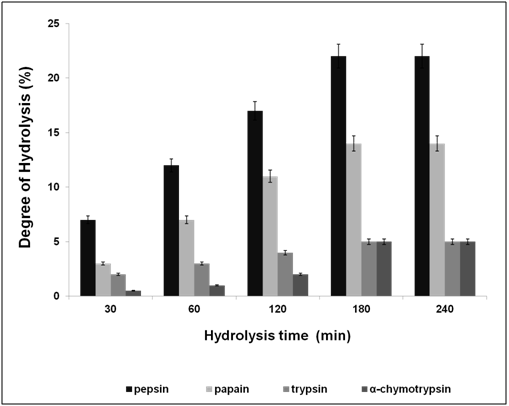
F. asafoetida root exudates protein was hydrolyzed enzymatically using simulated gastrointestinal fluid with enzymes (Papain, pepsin, trypsin, and α-chymotrypsin) which were optimized as shown in Table 1. The enzyme: substrate ratio (w/w) was 1:50 incubated at different time periods (30, 60, 90, 120 and 180 s) and then heated for 2 min at 100°C. The digested protein was separated in resolving 14% Laemmli, Glycine SDS-PAGE, followed by silver staining. Degree of hydrolysis was determined by measuring the nitrogen content soluble in 10% trichloroacetic acid as described previously [6].
Enzymes |
Buffer |
pH |
Temperature (°C) |
Papain |
50 mM Na2HPO4–NaH2PO4 |
6.5 |
37 |
Pepsin |
10 mM Tris-HCl |
2.0 |
37 |
Trypsin |
50 mM Tis-HCl |
7.8 |
37 |
α-chymotrypsin |
0.1 mM Tris-HCl |
7.8 |
37 |
Table 1 The optimum conditions of enzymatic hydrolysis of F. asafoetidaroot proteins with various enzymes
Separation of protein using Fast Protein Liquid Chromatography (FPLC)
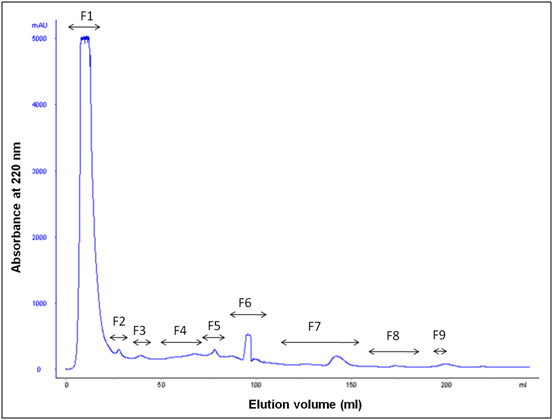
F. asafoetida root exudates hydrolyzed protein was loaded onto a Gel filtration column (Superdex 30 prep grade) connected to a fast protein liquid chromatography (FPLC). ÄKTApurifier™ chromatography system (ÄKTA purifier Frac-950 at a flow rate of 2 mL min-1) from GE Healthcare was used for fast and reliable separation of proteins. The column was equilibrated and eluted with 20 mM phosphate buffer (pH 7.2). The separation profile was monitored at wavelengths 280 and 220 nm using a UV detector. Each hydrolyzed protein was collected, lyophilized and stored at -20°C. These fractions were tested for antioxidant activity.
Antioxidant activity assay
ABTS radical-scavenging activity assay: The free radical scavenging capacity of hydrolyzed protein fractions was studied using the ABTS radical cation (ABTS+) decolorization assay, which is based on the reduction of ABTS+ radicals by antioxidants property of hydrolyzed protein. This assay was carried out in 96 well ELISA plates. ABTS 7 mM concentration was dissolved in deionized water. ABTS+ was produced by reacting ABTS solution with 2.45 mM potassium persulfate (final concentration) and allowing the mixture to stand in the dark condition at room temperature for 12-16 h, before use. In the experiments, the ABTS+ solution was diluted in deionized water or ethanol to 0.7 Å at 734 nm using Multiskan GO (Thermo Scientific, USA). An appropriate solvent blank reading was taken. The different concetration (2.5 µg to 30 µg) of hydrolyzed protein solution 20 µL was added to 190 µL of ABTS+ solution, follwed by incubation for 37°C at 30 min. The absorbance reading was taken at 734 nm and plotted against the linear standard curve between 20- 800 µM of Trolux soluion.17 All solutions were freshly prepared before expreiments and performed in triplicate. The percentage of inhibition of ABTS+ was calculated using the formula
Where, AB is the absorbance of the blank sample and AS is the absorbance of the sample.
Ferric reducing antioxidant power assay (FRAP): The FRAP assay was performed according to modified method of Thaipong et al.17 The stock solutions included 300 mM acetate buffer (3.1g C2H3NaO2.3H2O and 16mL C2H4O2, pH 3.6), 10mM TPTZ (2, 4, 6-tripyridyl-s-triazine) solution in 40mM HCl, and 20mM FeCl3.6H2O solution. The working solution was freshly prepared by mixing 25mL acetate buffer, 2.5mL TPTZ solution, and 2.5mL FeCl3.6H2O solution and heated at 37°C before use. The hydrolyzed protein fractions (25µl) were allowed to react with 175µL of the FRAP solution for 30min in the dark condition. This assay was carried out in 96 well ELISA plates. The absorbance reading of the colored product (ferrous tripyridyl triazine complex) were measured at 593nm using Multiskan GO (Thermo Scientific, USA) and plotted against the linear standard curve between 20- 800µM of Trolux soluion. All solutions were freshly prepared before expreiments and performed in triplicate.
Superoxide radical O2-scavenging activity assay: Superoxide radical scavenging activity was preformed according to a previously described method by Trisha et al.18 The reaction mixture consisted of 5.7 mL of 50 mM Tris-HCl, 1 mM EDTA (ethylene diamine tetraacetic acid) buffer, pH 8.6 (incubated at 25 °C for 20 min), 10µg of proteins from fractions and 0.1 mL of 5 mM pyrogallol solution (added in sequence). Pyrogallol was just used to initiate the reaction. The rate of pyrogallol antioxidation was measured at 320 nm using a spectrophotometer, which was its optical density and recorded every 30 s for 4 min. The capacity for scavenging superoxide anion radicals was calculated as
Where, AC is the change in absorbance per minute of the control solution containing pyrogallol and buffer, and AS is the change in absorbance per minute of the sample.
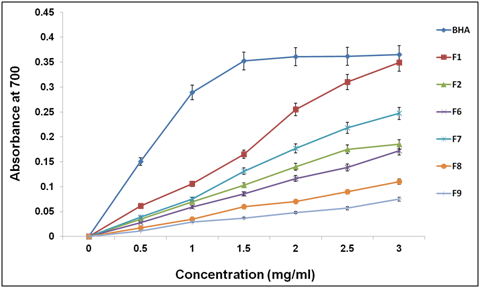
Reducing power assay: The reducing power of hydrolyzed protein fractions to reduce Fe (III) was measured according to the method of Ali Bougatef et al.19 In short, 0.5 mL of hydrolyzed protein fractions and 0.2 mL of 200 mM phosphate buffer (pH 6.6) were mixed with 50μL of 1 % K3Fe(CN)6. The mixture was incubated at 50 °C for 20 min and mixed with 0.1 mL of 10 % trichloroacetic acid (TCA) followed by centrifugation at 10000 rpm for 10 min. Further, 0.2 mL of the supernatant was collected and mixed with 0.2 mL of water, followed by 20 μL of 0.1% FeCl3. After 10 min of incubation at room temperature, the absorbance at 700 nm was measured for the resulting Prussian blue solution. The obtained results were compared with the BHA (butylated hydroxy anisole) standard. A higher absorbance value indicates greater reducing power.
Extraction and hydrolysis of F. asafoetida root exudates protein
After water extraction of F. asafoetida root exudates protein, it was quantified using Bradford’s method.20 Finally, lyophilized powder of 150 mg/g of protein was obtained. The molecular sizes of lyophilized proteins were determined using glycine SDS-PAGE as shown in Figure 2. As shown in Figure 2, the gel profile showed a lot of proteins including polypeptides and peptides in the molecular sizes in the range of 43 KDa to 3.5 KDa. Result showed that the major large protein molecules were above 10 kDa and minor group was small molecules less than 10 kDa, which are also called as peptides. F. asafoetida root exudates protein was hydrolyzed using in vitro gastrointestinal enzyme by varying the time period and enzyme: substrate (w/w) concentration. F. asafoetida root exudates proteins were separately hydrolyzed by papain, pepsin, trypsin, and α-chymotrypsin for the production of antioxidant peptides. The lyophilized F. asafoetida protein hydrolysate was dissolved in 20 mM sodium phosphate buffer (pH 7.2), and loaded onto a Superdex 30 column using fast protein liquid chromatography (FPLC). Fraction peaks were monitored for absorbance at 220 and 280 nm and each fraction were collected at rate of 2 mL/min. Figure 3 shows the elution profile of hydrolysate produced by pepsin digested protein. Totally 9 pooled fractions were collected based on peaks, lyophilized and tested for antioxidant activity. The gastric enzyme pepsin digested 26 kDaF. asafoetida protein into 14 kDa and 6.5 kDa accumulation and the maximum digestion was attained at 180 min. The enzyme Papain digested 26 kDa F. asafoetida protein into 3.5 kDa accumulation at 120 min. The pancreatic enzymes such as trypsin and α-chymotrypsin didn’t show any digestion at maximum time 180 min. The degree of hydrolysis for F. asafoetida root exudates protein was tested using the gastrointestinal enzymes. The protein degradation by proteolytic enzymes was estimated by assessing the degree of hydrolysis (DH) and it was observed that pepsin showed higher DH compared with other enzymes as shown in Figure 4. The protein degradation by proteolytic enzymes was observed to be 21.5 % and 13.2 % for pepsin and Papain. The other enzymes such as trypsin and α-chymotrypsin showed lower than 5 % degree of hydrolysis. As a result, enzyme pepsin was selected for further process. As shown in Figure 5, pepsin hydrolysates and crude protein antioxidant activity was compared to the natural antioxidant trolox and ascorbic acid. Results also depicted that pepsin hydrolysates antioxidant activity was comparable with the trolox antioxidant activity. Further, it was assumed that protein molecules are responsible for antioxidant agents, because the antioxidant activity against ABTS radicals scavenging activity increases significantly with the F. asafoetida hydrolysates concentration. In numerous literatures it has been reported that the highest antioxidant activity of proteins molecular weight was under 10 kDa.19,21-23

Antioxidant activity
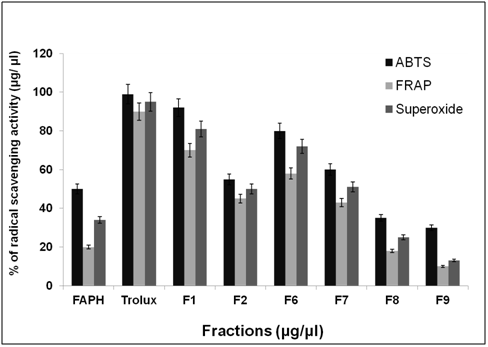
After purification by FPLC, nine fractions were collected and tested for antioxidant activity as shown in Figure 6. ABTS, FRAP and superoxide radical scavenging activity was used to estimate the antioxidant mechanism, due to the rapid reaction process (i.e. within 5 to 10 mins). The antioxidant activities of hydrolyzed factions were tested using ABTS, FRAP, Superoxide assay. The ABTS+ radical scavenging activity of pepsin digested F. asafoetida root exudates protein was tested. The relative antioxidant ability to scavenge the radical ABTS+ has been compared to the standards trolox and ascorbic acid, which is an excellent tool for determining the antioxidant activity. The result showed that there was an increase in the ABTS radical scavenging capacity of F. asafoetida root protein exudates up to a concentration of 25 µg/µl followed by comparatively low increase, with further increases in concentration. The result clearly depicted that ABTS+ assay for Fraction (F1, F2, F6) had maximum activity of 92, 75, 81% and F7, F8, F9 was < 60 % of activity. The other assay, FRAP showed F1, F2, F6 fraction had a maximum activity of 73, 60, 58 % and F7, F8, F9 had activity of < 40%. Finally, the reproducibility assay of superoxide scavenging activity showed that F1, F2, F3 had maximum activity of 81, 65, 72 % and F7, F8, F9 had activity of < 50%. From the result, Fraction 1 showed higher antioxidant properties and exhibited considerable scavenging potencies on ABTS+, FRAP, superoxide assays. Among three assays carried out in this study, ABTS showed comparatively enhanced activity than that of others. Another advantage of the ABTS was that extracts reacted rapidly with ABTS. Among the fractions, fraction 1 showed the highest peak and higher antioxidant activity compared to other fractions in three different antioxidant assays. As the response of antioxidants depends on many factors, the antioxidant activity of the hydrolysates was characterized using different assays based on different antioxidant mechanisms. The ABTS radical cation scavenging assay is applicable to both lipophilic and hydrophilic compounds and has been widely used to assess antioxidant activity.24 ABTS assay is an excellent tool for determining the antioxidant activity of hydrogen-donating antioxidants (scavengers of aqueous phase radicals) and of chain breaking antioxidants (scavenger of lipid peroxyl radicals).25
The antioxidant activity of hydrolysates is mainly influenced by their amino acid sequence, which depends on the protease specificity.26-29 It is reported that both large peptides and short ones can protect from oxidation by various mechanisms, including metal Chelation and formation of oil droplets around membranes.29 The ferric reducing antioxidant power, generally measures the antioxidant effect of any substance in the reaction medium as its reducing ability. At low pH, when a ferric tripyridyl triazine complex is reduced to the ferrous form, an intense blue color with an absorption maximum at 593 nm develops. The reaction is nonspecific and any half-reaction which has a less-positive redox potential, under reaction conditions, than the FeIII/FeII-TPTZ half-reaction will drive FeIII-TPTZ reduction. Test conditions favor the reduction of the complex and thereby, color development, provided that a reductant (antioxidant) is present.30 Superoxide radical is known to be very harmful to cellular components as a precursor of more reactive oxidative species, such as single oxygen and hydroxyl radicals.31 Furthermore, superoxide radical is considered to play an important role in the peroxidation of lipids. The reducing power assay is often used to evaluate the ability of antioxidant to donate electron.32 Further, detailed studies of the amino acid sequence responsible for the antioxidant activity have to be elucidated.
Reducing power assay
The reducing power assay of hydrolyzed protein was investigated following the method reported by Zhu et al.22 and the results are shown in Figure 7. To measure the reductive ability of protein hydrolysates, different fraction were studied for the reduction of Fe3+ to Fe2+. The reducing power of fraction (1) was excellent, which increased steadily with increase in the concentration of samples. At 3 mg, the power of reducing was higher than 0.38. Thus, this data supports the antioxidant activity of fractions.
The enzymatic hydrolysis of F. asafoetida through the action of pepsin provided a high proportion of peptides in the range of 10 kDa - 6.5 kDa. The hydrolysis degree of pepsin digested F. asafoetida showed maximum (22 %) hydrolysis for 180 min. After purification using FPLC, the hydrolysates showed more antioxidant activity in three different radical scavenging assays. The ABTS, FRAP, and superoxide radical assays gave comparable results for the antioxidant activity measured in F. asafoetida root protein exudates. The ABTS technique was simple, rapidly performed and showed high reproducible activity. Therefore, it would be an appropriate technique for determining antioxidant in F. asafoetida root protein exudates. These hydrolysates can be used to formulate functional food ingredient in pharmaceutical and nutraceuticals. However, further detailed studies on its peptide fractions with regard to amino acid sequence, antioxidant activity in vivo and the different antioxidant pathway mechanisms are needed.
Author declares that there is no conflict of interest.

©2016 Prabaharan, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.