International Journal of
eISSN: 2573-2889


Research Article Volume 4 Issue 2
1Department of Pathology, National Institute of Nutrition (ICMR), India
2Department of Pathology, ESIC Medical College, India
3Department of Pathology & Microbiology, National Institute of Nutrition (ICMR), India
4Department of Biochemistry, National Institute of Nutrition (ICMR), India
Correspondence: Suresh Challa, Scientist E, & Deputy Director, Department of Biochemistry, National Institute of Nutrition (ICMR), Tarnaka, Hyderabad, 500007, India
Received: January 31, 2019 | Published: April 2, 2019
Citation: Thathapudi S, Erukambattu JS, Putcha UK, et al. Multifactor dimensionality reduction analysis for detecting SNP-SNP, SNP-environment interactions associated with polycystic ovarian syndrome among South Indian women. Int J Mol Biol Open Access. 2019;4(2):59?65. DOI: 10.15406/ijmboa.2019.04.00098
Background: Polycystic Ovarian Syndrome (PCOS) is the most common cause of female anovulatory infertility, with a prevalence of 8-10% among women of reproductive age, characterized by clinical or biochemical androgen excess, ovulatory dysfunction and polycystic ovaries. It is a multifactorial disorder which is determined by the interaction of multiple genetic and environmental factors. Many candidate genes have been proposed as important contributors to PCOS. One of the greatest challenges facing human geniticists is the identification and characterization of susceptibility genes for common complex multifactorial human diseases. This challenge is partly due to the limitations of parametric-statistical methods for detection of gene effects that are dependent completely or partially on interactions with other genes with environmental factors. Multifactor-dimensionality reduction (MDR) has power to identify interactions among two or more loci in relatively small samples, and reduces dimentionality of multilocus information and improves identification of polymorphism combinations associated with disease risk.
Aim: Our aim is to identify the effect of gene – gene, and gene- environmental interactions associated with PCOS disease risk using MDR.
Materials and methods: Prospective genetic case-control study, involving 204 women with PCOS and 204 healthy, sex and age matched controls. Anthropometric and biochemical profile were taken in a well-designed proforma. Isolation of DNA by salting out method and genotype analysis was done for all the study population using PCR-RFLP. MDR analysis of SNPs and SNP–environmental interactions were done.
Results: MDR statistical version (MDR_3.0.2) showed the strong interaction between Calpain-10-FSHR, and TNF-α-IGF2. Moderate interaction is noticed between LHCGR, TNF-α/IGF2, and weak interaction between LHCGR and FSHR/CAPN-10. Correlation is seen between TNF-α and IGF2, synergistic interaction seen between FSHR and Calpain-10, whereas LHCGR is independently interacting with the disease. SNP–environmental interactions also revealed impact of individual factor and their combination with SNP has shown significant effect in disease risk.
Conclusion: SNP-SNP, SNP-Environmental factors interaction analysis using MDR software revealed the role of various genes and environmental factors in combination are increasing the disease risk effectively, and helps in better understanding of the reproductive and metabolic abnormalities and management of the PCOS.
Keywords: polycystic ovary syndrome, multifactor dimensionality reduction, SNP
Polycystic ovarian syndrome (PCOS) is the leading cause of menstrual irregularities and anovulatory infertility which is now essentially known as androgen excess disorder with varying degrees of reproductive and metabolic abnormalities determined by the interaction of multiple genetic and environmental factors. We focused on the candidate gene analysis of PCOS in the South Indian women by analyzing a panel of five candidate genes (Tumor necrosis factor alpha A, Insulin growth factor 2, Calpain 10, Follicular stimulating hormone receptor and Leutinising hormone G protein coupled receptor) involved in insulin action and secretion, gonadotropin action and regulation and hyperandrogenism. The genetic association of the above candidate genes with PCOS susceptibility were studied.1–5 However in the case of complex genetic disorders, it is usually observed that inspite of a small effect of an individual SNP, the genetic effects of combinations of functionally relevant SNPs may synergistically contribute to increased disease risk, One of the biggest challenges in human genetics is identifying polymorphisms, or mutations, that increases the disease risk. This challenge is partly due to the limitations of parametric statistical methods (i.e., those in which a hypothesis about the value of a statistical parameter is made) for detection of gene effects that are dependent completely or partially on interactions with other genes6 and with environmental exposures.7 Multifactor dimensionality reduction (MDR) was developed for detecting gene-gene and gene-environment interactions in case-control studies with relatively small sample size.
Ethics Statement
This study was approved by the institutional ethical committee (Vasavi hospital and research centre) and informed written consent was obtained from all subjects. In this prospective case-control study, we included 204 PCOS patients from Anu’s fertility center, Somajiguda, Hyderabad, India from July 2011 to January 2013.
Inclusion criteria
Subjects were ranged in age from 17 to 35 years and were diagnosed using the 2006-Androgen Excess Society (AES) Criteria. Hyperandrogenism, clinical or biochemical and either Oligo-anovulation or Polycystic ovarian morphology.8
Exclusion criteria
Women excluded from the study were those with inherited disorders like congenital adrenal hyperplasia, androgen secreting neoplasms, androgenic/anabolic drug use or abuse, Cushing syndrome, syndromes of severe insulin resistance, thyroid dysfunction and hyperprolactinemia. We have recruited 204 controls from a tertiary care hospital, Kamineni academy of medical sciences and research center, LB Nagar, Hyderabad. Controls ranged from 17-35 years and did not show hirsutism, acne or male-type alopecia and had regular menstrual cycles and none of them satisfied any of the AES-2006 criteria. All the control subjects also underwent an ultrasonographic examination, and women who had any pathologic findings like polycystic ovaries were excluded from the study.1–5
Collection of blood samples, DNA isolation, and SNP Analysis by using PCR-RFLP
Two milliliters of peripheral blood was collected in EDTA for DNA isolation according to the method routinely used in our laboratory, and 5 ml of blood in plain vial for serum preparation from all the patients and controls along with clinical data, personal history and family history. We carried out PCR - RFLP to screen the polymorphisms of TNFα, IGF2, CAPN-10, FSHR, LHCGR genes.1–5
We framed our analysis of genetic polymorphisms in four steps: 1.SNP-SNP interaction analysis using different combinations among the candidate gene panel. 2. SNP-Environment interaction Analysis. 2.a. SNP-Clinical interaction analysis between each genetic polymorphic locus and clinical factors considered as categorical variables. 2.b. SNP- Biochemical interaction analysis between each genetic polymorphic locus and Biochemical factors considered as categorical variables. 2.c. SNP-Hormonal interaction analysis between each genetic polymorphic locus and hormonal factors considered as categorical variables. TNFα, IGF2, CAPN-10, FSHR, LHCGR genes were involved in SNP-SNP, SNP-Environment (SNP-Clinical, SNP-Biochemical & SNP-Hormonal) interaction analysis.
For SNP-SNP interaction analysis, we considered five genes (TNFα, IGF2, CAPN-10, FSHR and LHCGR), for gene-clinical: six factors (acne, hirsutism, central obesity, fertility, menarche age, menstrual period), for SNP-Biochemical: nine factors (fasting glucose, fasting Insulin, HOMA score, LH, FSH, LH/FSH, HDL, Triglycerides, TNF serum levels), for SNP-Hormonal: four factors (free testosterone, total testosterone, androstenedione, DHEA) were considered.
Statistical analysis
A p-value of <0.05 was considered statistically significant. Multifactor dimensionality reduction (MDR) software was used for carrying out SNP-SNP and SNP-Environment interaction analysis.
Anthropometric, biochemical and hormonal findings showed significant differences between PCOS and Controls (Table 1–4). Increased BMI and HOMA is associated with IGF2, FSHR and LHCGR gene polymorphism. Increased LH, LH/FSH and DHEA is associated with FSHR and LHCGR gene polymorphism. Increased FTS is associated with IGF2 and FSHR gene polymorphism. The variants of TNFα C850T, Apa1 IGF2 A820G, FSHR Ser680Asn and LHCGR A312G showed 5.7, 7.6, 1.98, & 3.36 folds risk of developing PCOS in our population. CAPN-10 UCSNP-43 did not show any risk association with PCOS in our population.
Parameter |
Patients(n=204) |
Controls(n=204) |
P value |
Age(years) |
28±3.6 |
28± 5.1 |
1 |
Body mass index (kg/m2) |
27.12±4.93 |
23.4±3.2 |
<0.0001 |
Waist circumference (inches) |
37±4.3 |
30.36±3.3 |
<0.0001 |
Hip circumference(inches) |
39.4±4.1 |
38.11±3.7 |
0.0008 |
Waist to Hip ratio |
0.93±0.04 |
0.79±0.05 |
<0.0001 |
Table 1 Comparison of anthropometric parameters between PCOS and controls using mean and standard deviation
Parameter |
Patients (n=204) |
Controls (n=204) |
P value |
Fasting glucose (gm/dl) |
88±8.6 |
86.85±7.1 |
0.0678 |
Fasting insulin (Uu/ML) |
16.94±7.26 |
6.66±3.19 |
<0.0001 |
Homa Score |
3.73±3.8 |
1.44±0.75 |
<0.0001 |
Lh (IU/L) |
11.97±6.08 |
5.5±3.8 |
<0.0001 |
Fsh (IU/L) |
5.48±1.98 |
7.9±5.4 |
0.0002 |
LH/FSH |
2.62±1.2 |
1.5±1.2 |
<0.0001 |
Cholesterol (mg/dl) |
161.57±30 |
162.7±33 |
0.8904 |
Hdl (mg/dl) |
40.21 ±10.21 |
45±13.4 |
<0.0001 |
Triglycerides (mg/dl) |
126.14±45.2 |
97±49 |
<0.0001 |
Tnf-α (pg/ml) |
13.24±10.6 |
5.6±3.86 |
<0.0001 |
Table 2 Comparison of biochemical parameters between PCOS and controls using mean and standard deviation
Parameter |
Patients (n=204) |
Controls (n=204) |
P value |
Total testosterone (ng/ml) |
5.8±4.319 |
1.32±1.05 |
<0.0001 |
Free testosterone (pg/ml) |
8.39±6.69 |
2.6±1.4 |
<0.0001 |
Androstenedione (ng/ml) |
2.41±1.5 |
1.046±0.68 |
<0.0001 |
Dihydroxyepiandrostenedione (DHEA) (ng/ml) |
6.22±5.6 |
1.9±0.9 |
<0.0001 |
Table 3 Comparison of Hormonal parameters between PCOS and controls using mean and standard deviation
Gene |
rs ID |
Location |
Allele |
PCOS (N=204) |
Controls (N=204) |
Odds ratio (95% C.I) |
P-value |
TNF-α |
rs 1799724 |
Exon |
C |
382 |
302 |
5.1569 |
P<0.0001 |
C850T |
T |
26 |
106 |
(3.27-8.12) |
|||
IGF2 ApaI |
rs 680 |
3’UTR |
A |
244 |
375 |
7.639 |
P<0.0001 |
A820G |
G |
164 |
33 |
(5.083-11.47) |
|||
Calpain 10 |
rs 3792267 |
Intron 3 |
G |
347 |
328 |
0.7207 |
P=0.0793 |
UCSNP-43 |
A |
61 |
80 |
(0.5-1.039) |
|||
FSHR |
rs6166 |
Exon 10 |
G |
161 |
230 |
1.98 |
P<0.0001 |
Ser680Asn |
A |
247 |
178 |
(1.5-2.6) |
|||
LHCGR |
Rs2293275 |
Exon 10 |
G |
242 |
199 |
1.5311 |
P=0.0026 |
S312N |
|
|
A |
166 |
209 |
(1.16-2.01) |
|
Table 4 Summary table for significant observation in genotype/allele frequency analysis for individual genes
SNP-SNP, SNP-environmental, SNP-biochemical and SNP-hormonal interaction analysis
The multifactorial dimensionality reduction analysis revealed significant contribution of four allelic variants i.e. TNF-α, IGF2, FSHR, and LHCGR in modulating the PCOS risk (Figure 1). CAPN-10 was the only polymorphism evaluated which did not show any association. The shorter the line connecting to attributes the stronger the interaction. The color of the line indicates the type of interaction. Red suggests that there is synergistic relationship (epistasis), Yellow suggests independence, Green and blue suggests redundancy or correlation (Figure 2). When the interaction of five gene polymorphisms was evaluated using MDR statistical version (MDR_3.0.2) showed the strong interaction between Calpain-10-FSHR, and TNF-α-IGF2. Moderate interaction is noticed between LHCGR, TNF-α/IGF2, and weak interaction between LHCGR and FSHR/CAPN-10. Correlation is seen between TNF-α and IGF2, synergistic interaction seen between FSHR and Calpain-10, whereas LHCGR is independently interacting with the disease. The interaction graph presents that A820 A/G IGF2 Apa1 SNP (24.28%), 850 C/T TNF-α (12.24%), 680 G/A FSHR (4.36%), 312 A/G LHCGR (4.10%), UCSNP-43 G/A CAPN-10 (0.71%) polymorphism have independent effects for developing PCOS (Figure 3) (Table 5). There were found independent effects with 0.40% between menarche age and 312A/G LHCGR polymorphism and also found that central obesity, hirsutism, menstrual period, acne, fertility, menarche age, have independent effects for PCOS developing 85.72% entropy for central obesity, followed by 78.81 %,73.67%, 72.87%, 63.93%, 4.68% of entropy. TNF-α, IGF2, CAPN-10 and FSHR showed correlation effects with negative entropy (Figure 4) (Table 6). The interaction graph showed combination of TNF-α- fasting glucose, IGF2-fasting glucose, IGF2-HDL, IGF2-FSH, CAPN10-fasting glucose, CAPN10-HDL, CAPN10-fasting insulin, CAPN10-LH/FSH, FSHR-LH, FSHR-HDL, FSHR-fasting insulin, LHCGR-HOMA, LHCGR-LH, LHCGR-TG, LHCGR-TNFα showed synergistic effect with 0.46% of entropy followed by 0.47%, 1.02%, 0.40%, 0.53%, 0.63%, 0.42%, 0.09%, 0.08%, 0.35%, 0.38%, 0.29%, 0.17%, 0.62%, 0.47%, 0.29% of entropy. It was also found that the combination of TNF-α, IGF2, CALP-10, FSHR with serum TNF-α showed correlation with negative entropy (Table 7). The interaction graph presents that Androstenedione, FTS, DHEA, TTS have independent effects for PCOS developing with 27.50% of entropy for androstenedione, followed by 26.99%, 16.23%, 7.93% of entropy. It was found that combination of FSHR-TTS, CALP10-AND, CALP10-DHEA, CALP10-TTS, and CALP10-FTS are risk factors for PCOS, showed independent effects with 0.97%, followed by 1.09%, 0.41%, 0.34% and 0.03% of entropy (Figure 5) (Table 8).
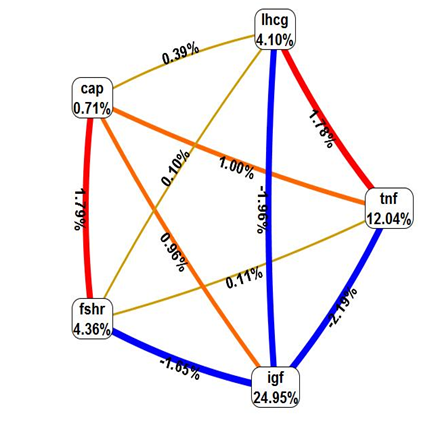
Figure 1 Circle graph representing gene-gene interaction of insulin resistance and hypothalamo-pituitary-gonadotropin axis pathway in PCOS.

Figure 2 Dendogram representing gene-gene interaction of insulin resistance and hypothalamo-pituitary-gonadotropin axis pathway.
Loci combinations |
SNP combinations |
Balanced accuracy values |
CV consistency |
p-value |
Two loci |
TNF, IGF2 |
0.7868 |
10 |
0.0002 |
TNF, CAPN10 |
0.6667 |
10 |
0.0195 |
|
TNF, FSHR |
0.7108 |
10 |
0.0068 |
|
TNF, LHCGR |
0.6691 |
10 |
0.018 |
|
IGF2, CAPN10 |
0.7794 |
10 |
0.003 |
|
IGF2, FSHR |
0.7794 |
10 |
0.003 |
|
IGF2, LHCGR |
0.7598 |
6 |
0.0009 |
|
CAPN10, FSHR |
0.5711 |
8 |
0.3638 |
|
CAPN10, LHCGR |
0.5711 |
10 |
0.2862 |
|
FSHR, LHCGR |
0.5613 |
10 |
0.4335 |
|
Three loci |
TNF, IGF2, CAPN10 |
0.7721 |
10 |
0.0004 |
TNF, IGF2, FSHR |
0.777 |
10 |
0.0003 |
|
TNF, IGF2, LHCGR |
0.8064 |
10 |
0.0002 |
|
IGF2, CAPN10, FSHR |
0.7696 |
10 |
0.0005 |
|
IGF2, CAPN10, LHCGR |
0.7574 |
10 |
0.001 |
|
CAPN10, FSHR, LHCGR |
0.576 |
10 |
0.3304 |
|
CAPN10, FSHR, TNF |
0.6618 |
10 |
0.0361 |
|
TNF, FSHR, LHCGR |
0.6936 |
10 |
0.0108 |
|
IGF2, FSHR, LHCGR |
0.7574 |
9 |
0.001 |
|
Four loci |
TNF, IGF, CAPN10, FSHR |
0.7353 |
10 |
0.0026 |
TNF, CAPN10, FSHR,LHCGR |
0.625 |
10 |
0.1056 |
|
LHCGR,CAPN10,FSHR,IGF2 |
0.7206 |
10 |
0.0048 |
|
TNF,IGF2,FSHR,LHCGR |
0.7574 |
7 |
0.001 |
|
TNF,IGF2,CAPN10,LHCGR |
0.7868 |
10 |
0.0002 |
|
Five loci |
IGF2,TNF,CAPN10,FSHR, LHCGR |
0.7255 |
10 |
0.0039 |
Table 5 SNP- SNP interactions in each loci combination category
Loci combinations |
SNP combinations |
Balanced accuracy values |
CV consistency |
p-value |
6 Loci |
FSHR, LHCGR, CAPN10, TNF,IGF2, Acne |
0.9412 |
10 |
<0.0001 |
FSHR,LHCGR,CAPN10,TNF,IGF2,Central obesity |
0.9755 |
10 |
<0.0001 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Hirsutism |
0.9583 |
10 |
<0.0001 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Fertility |
0.9289 |
10 |
<0.0001 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Menarche age |
0.7525 |
10 |
0.0012 |
|
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Menustrual period |
0.9436 |
10 |
<0.0001 |
Table 6 Gene- Environment (clinical) interactions
Loci combinations |
SNP combinations |
Balanced accuracy values |
CV consistency |
p-value |
6 Loci |
FSHR, LHCGR, CAPN10, TNF, IGF2, Fasting Insulin |
0.799 |
10 |
0.0001 |
FSHR,LHCGR,CAPN10,TNF,IGF2,Homa Score |
0.7623 |
5 |
0.0008 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,LH |
0.799 |
10 |
0.0001 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,LH/FSH |
0.7181 |
10 |
0.0047 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Triglycerides |
0.7181 |
10 |
0.005 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,HDL |
0.7377 |
10 |
0.0023 |
|
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Serum TNF |
0.7328 |
10 |
0.0009 |
Table 7 Gene- Environment (Biochemical) interactions
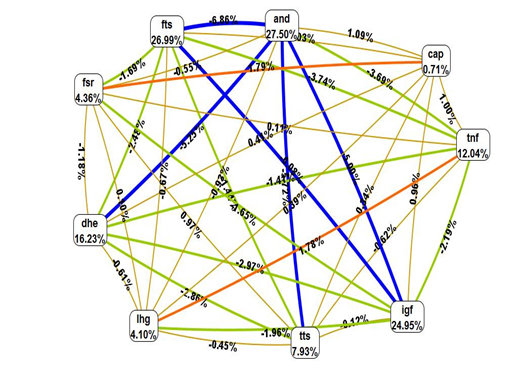
Figure 5 Circle graph representing gene–androgen levels interaction of five gene polymorphisms in PCOS as compared to controls.
Loci combinations |
SNP combinations |
Balanced accuracy values |
CV consistency |
p-value |
6 Loci |
FSHR,LHCGR,CAPN10,TNF,IGF2, Total Testosterone |
0.6569 |
10 |
0.0335 |
FSHR,LHCGR,CAPN10,TNF,IGF2,Free Testosterone |
0.723 |
10 |
0.0006 |
|
FSHR,LHCGR,CAPN10,TNF,IGF2,DHEA |
0.6446 |
10 |
0.0086 |
|
|
FSHR,LHCGR,CAPN10,TNF,IGF2,Androstenodian |
0.7475 |
10 |
0.0003 |
Table 8 Gene- Environment (Hormonal) interactions
Complex multifactorial disorders are manifested as a result of interactions between multiple genetic and environmental factors9,10 because the effect of any single genetic variation will likely be dependent on other genetic variations (gene-gene interaction or epistasis) and environmental factors (gene-environment interaction). MDR addresses concerns about inaccurate parameter estimates and low power for identifying interactions in relatively small sample sizes. MDR is a nonparametric and genetic model-free approach. In MDR, genotypes are pooled into high risk and low risk groups, effectively reducing the dimensionality of the genotype predictors from N dimensions to one dimension. The new one –dimensional multi locus genotype variable is evaluated for its ability to classify and predict disease status using cross-validation and permutation testing. In the dendrogram, color of the line indicates the type of interaction. Red and Orange suggest there is a synergistic relationship (i.e. epistasis). Yellow suggests independence. Green and Blue suggest redundancy or correlation The shorter the line connecting two attributes the stronger the interaction (Epistasis blog). Cordell reviewed, multifactor dimensionality reduction has emerged as one important new method for detecting statistical epistasis in genetic association studies.11 MDR was a nonparametric method and genetic model- free data mining and machine learning strategy for identifying combinations of discrete genetic and environmental factors.12–18 PCOS is a multifactorial disorder which is determined by the interaction of multiple genetic and environmental factors. Many candidate genes have been proposed as important contributors to PCOS. We focused on the candidate gene analysis of PCOS in the South Indian women by analyzing a panel of five candidate genes (Tumor necrosis factor alpha A, Insulin growth factor 2, Calpain 10, Follicular stimulating hormone receptor and Leutinising hormone G protein coupled receptor) involved in insulin action and secretion, gonadotropin action and regulation and hyperandrogenism. The genetic association of the above candidate genes with PCOS susceptibility were studied.1–5 The variants of TNFα C850T, Apa1 IGF2 A820G, FSHR Ser680Asn and LHCGR A312G showed 5.7, 7.6, 1.98, & 3.36 folds risk of developing PCOS in our population. CAPN-10 UCSNP-43 did not show any risk association with PCOS in our population. Although the candidate genes and the underlying biological pathways analyzed in this study have been previously implicated in the etiology of PCOS, their SNP-SNP interactions, SNP-environment interactions have not been described before. Therefore to identify the complex biological relationships between the molecular pathological pathways leading to PCOS, we attempted to understand the epistasis phenomenon involved in PCOS etiology through SNP-SNP interaction, SNP- environment interaction analysis. In the individual gene analysis, four out of five genes indicated significant associative patterns with PCOS, and Calpain-10 did not show any contribution, but it showed a positive interaction with FSHR and TNF-α polymorphism. The first genome-wide association study on PCOS was conducted among the Han-Chinese population, which led to the identification of a more specific genomic region that may contain the candidate genes specific to the Han-Chinese population.19
To the best of our knowledge, ours is the first study to show SNP-SNP and SNP-Environment interactions incorporated in insulin action and secretion, gonadotrophin action and regulation and hyperandrogenism pathways. The multifactorial dimensionality reduction analysis revealed significant contribution of SNP-SNP, SNP-Environmental interactions in modulating the PCOS risk. MDR statistical version (MDR_3.0.2) showed the strong interaction between Calpain-10-FSHR, and TNF-α-IGF2. Moderate interaction between LHCGR, TNF-α/IGF2, and weak interaction between LHCGR and FSHR/CAPN-10. Correlation is seen between TNF-α and IGF2, synergistic interaction seen between FSHR and Calpain-10, whereas LHCGR is independently interacting with the disease. The interaction graph presents independent effects with A820 A/G IGF2 Apa1 SNP (24.28%), 850 C/T TNF-α (12.24%), 680 G/A FSHR (4.36%), 312 A/G LHCGR (4.10%), UCSNP-43 G/A CAPN-10 (0.71%) polymorphism have independent effects for developing PCOS. For a multifactorial disorder like PCOS, it is important to obtain the genetic and environment risk factors resulted by their joint interactions, in understanding PCOS pathogenesis, and its management.
SNP-SNP, SNP-Environmental factors interaction analysis using MDR software revealed the role of various genes and environmental factors in combination are increasing the disease risk effectively, and helps in better understanding of the reproductive and metabolic abnormalities and management of the PCOS.
None.
The author declares there is no conflicts of interest.

©2019 Thathapudi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.