International Journal of
eISSN: 2573-2889


Research Article Volume 3 Issue 6
Independent researcher and analyst, Computational Biology, India
Correspondence: Anubha Dubey, Independent researcher and analyst, Computational Biology, Gayatri Nagar atni, MP, India
Received: November 26, 2018 | Published: December 31, 2018
Citation: Dubey A. Machine learning model for analysis of critically important antimicrobials for human medicine. Int J Mol Biol Open Access. 2018;3(6):287-293. DOI: 10.15406/ijmboa.2018.03.00089
With the development of antimicrobials, microbes have adapted and become resistant to previous antimicrobial agents. Hence WHO recommended complete list of critically important antimicrobials, highly important and important antimicrobials. So there is a need to classify critically important antimicrobials for human medicine so these can be used only for humans. Therefore machine learning model is developed in this paper to classify critically important antimicrobials based on their amino acid composition with great accuracy.
Keywords: antimicrobials, WHO, machine learning, amino acid composition
The science and practice of the diagnosis, treatment, and prevention of disease is called medicine. Properties of medicine are maintenance and restoration of health by the preventing and treating the ill effects. They are responsible for killing or slow down the microbial growth. Any kind of bacteria, viruses etc that are not visible to naked eyes are called micro-organisms or microbes. Some category of microbe is available in Table 1.
Microbe |
Example |
Type of infection caused |
Bacteria |
Staphylococcus aureus, etc |
Some staph infections |
Virus |
Influenza |
Flu |
Fungi |
Candida albicans, etc |
Yeast infections |
Parasites |
Plasmodium falciparum, etc |
Malaria |
Table 1 Variety of microbes with example and their infection
For treating human diseases different variety of antimicrobial classes are used. These antimicrobials if used regularly develop resistance called antimicrobial resistance. And the genes responsible for resistance are called anti microbial resistance. For example, the ndm-1 gene encodes resistance to the carbapenem family was first discovered in Klebsiella pneumonia that was isolated from an infected person.1 Most of the AMR are hazardous to human health. Characteristics by which antimicrobials are classified are as follows:
Characteristic 1 (C1): The class that treat serious ill effects caused by bacteria in people.
Characteristic 2 (C2): The action of antimicrobials include:(a) Bacteria that transmitted to humans from nonhuman sources, (b) Bacteria that may acquire genes for resistance from sources other than humans.
Antimicrobials vs antibiotics
The preventive measure in form of medicine are called antibiotics which work against bacteria and treat bacterial infections. When bacteria change their forms in response to the repeated use of antibiotics develops antibiotic resistance. Broadly antimicrobial resistance to drugs to treat infections caused by other microbes such as parasites (e.g. malaria), viruses (e.g. HIV) and fungi (e.g. Candida). Hence Antimicrobials are one of few alternatives for the treatment of serious bacterial infections in humans that occupies an important place in human medicine. Serious infections are likely to result in significant morbidity or mortality if left untreated. Multidrug resistance is also the outcomes of disease which relate to the site of infection e.g. pneumonia, meningitis or the host e.g. infant, immunosuppressant. The use of such antibacterial agents is preserved, as loss of efficacy in these drugs due to the emergence of resistance leads to significant impact on human health, especially for people with life-threatening infections. These are the alternatives for the treatment of serious bacterial infections in human that play an important role in human medicine. If infections left untreated there would be significant morbidity or mortality. Sometimes multidrug resistance would also occur like pneumonia, meningitis etc. The antimicrobial agents that used to treat diseases caused by bacteria are transmitted to humans from non-human sources i.e. water, food, environment or animal. These are considered as highly important antimicrobials because such infections are most amenable to risk management. Nonhuman sources and the bacteria causing human diseases are linked. Such example includes non-typhoidal salmonella, campylobacter spp. E. coli etc. This is called commensalism. The commensalisms themselves may also be pathogenic in immuno suppressed hosts. The transfer of their genes shows the transmission of AMR. Interpretation of categorization of antimicrobial class:
Critically important: Antimicrobial classes which meet both C1 and C2 are termed critically important for human medicine.
Highly important: Antimicrobial classes which meet either C1 or C2 are termed highly important for human medicine.
Important: Antimicrobial classes used in humans which meet neither C1 nor C2 are termed important for human medicine. The list below is meant to show examples of members of each class of drugs. All drugs that are listed in a given class have not necessarily been proven safe and effective for the diseases.2
There are many antimicrobials like Aminoglycocides, ansamycins, carbapenems and other penems, Cephalosporins, Glycopeptides, Glycylcyclines, lipopeptides, Macrolids and ketolids, monobactrum, Oxazolidinones, Penicillins, Phosphonic acid derivatives, Polymyxins, Quinolones, sulfones, Tetracyclines, Nitrofuratoins, etc are classified according to their mode of action and above explained three categories. All the details of these antimicrobials are explained in Table 2 which also describes their significance of treating disease and their causative organism respectively.
|
Antimicrobial class |
Example of drugs |
Mode of action |
Causative organism |
Treating disease |
References |
||
|
|
Critically important antimicrobials |
||||||
|
|
Aminoglycosides |
Gentamicin |
Irreversibly bind 30S ribosomal proteins (bactericidal) |
P. aeruginosa Gram negative bacteria |
Bone infections, endocarditis, pelvic inflammatory disease, meningitis, pneumonia, urinary tract infections |
||
|
|
Ansamycins |
Rifampicin |
DNA directed RNA polymerase |
Mycobacterium tuberculosis, Mycobactterium Kansasii |
Tuberculosis, mycobacterium avium complex, leprosy, and Legionnaire's disease |
||
|
|
Carbapenems and other penems |
Meropenem |
Inhibition of peptidoglycan synthesis (bactericidal) |
Many Gram-positive and Gram-negative bacteria (including Pseudomonas) and anaerobic bacteria. |
Meningitis, intra-abdominal infection, pneumonia, sepsis, and anthrax. |
||
|
|
Cephalosporins (3rd,4th and 5th generation) |
Ceftriaxone, cefepime, ceftaroline |
Cell wall synthesis |
Gram positive and gram negative bacteria i.e. , H. influenzae, and susceptible E. coli, Klebsiella, and penicillin-resistant N. gonorrhoeae |
Typhoid fever |
||
|
|
Glycopeptides |
Vancomycin |
Disrupts peptidoglycan cross-linkage |
Gram-negative bacteria Enterococci, Clostridium difficile |
Skin infections, bloodstream infections, endocarditis, bone and joint infections, |
||
|
|
Glycylcyclines |
Tigecycline |
Gram positive bacteria penicillin-resistant Streptococcus pneumoniae, methicillinresistant Staphylococcus aureus (MRSA) and Staphylococcus epidermidis (MRSE), and vancomycin-resistant Enterococcus (VRE) |
Neusea, vomiting,diarrhoea |
|||
|
|
Lipopeptides |
Daptomycin |
Cytoplasmic membrane structure |
S. aureus |
Skin and skin structure infections |
||
|
|
Macrolides and ketolides |
Erythromycin, Telethromycin |
Protein synthesis (50 s inhibitor) |
Erm encoded methylases in S. aureus |
Respiratory tract infections. |
||
|
|
Monobactrum |
Aztreonam |
Cell wall synthesis |
Gram-negative bacteria such as Pseudomonas aeruginosa |
Bone infections, endometritis, intra abdominal infections, pneumonia, urinary tract infections, and sepsis. |
||
|
|
Oxazolidinones |
Linezolid |
Protein synthesis inhibitor |
E. facium and S. aureus |
Infection of skin and pneumonia |
31. |
|
|
|
Penicillins (natural, aminopenicillins, |
Ampicillin |
Cell wall synthesis |
Group B streptococcal infection in newborn |
respiratory tract infections, urinary tract infections, meningitis, salmonellosis, and endocarditis |
||
|
|
Phosphonic acid derivatives |
Fosfomycin |
Bacterial cell wall biogenesis |
Proteus spp., Enterobacter spp., Citrobacter spp., Serratiamarcescens and Salmonella enterica E. faecalis, E. coli |
Sepsis, urinary tract infections |
||
|
|
Polymyxins |
Colistin |
cytoplasmic membrane structure |
Pseudomonas aeruginosa, Klebsiella pneumoniae and Acinetobacter. |
Kidney infections |
||
|
|
Quinolones |
Ciprofloxacin |
DNA gyrase |
Kill growth of bacteria |
chest infections, urine infections, prostatitis, infections of the digestive system, bone and joint infections, and some sexually transmitted infections. |
||
|
|
Drugs used solely to treat tuberculosis or other mycobacterial diseases |
Isoniazid |
Isoniazid is a prodrug and must be activated by a bacterial catalase-peroxidase enzyme |
or atypical types of mycobacteria, such as M. avium, M. kansasii, and M. xenopi. |
Tuberculosis |
||
|
|
Highly important antimicrobials |
|
|
|
|
||
|
|
Amidinopenicillins |
Mecillinam |
Cell wall synthesis |
Escherichia coli. most pathogenic Gram-negative bacteria, except Pseudomonas, k; paeruginosa and some species of Proteus. |
Urinary tract infections, and has also been used to treat typhoid and paratyphoid fever. |
||
|
|
Amphenicols |
Chloramphenicol |
Cytoplasmic membrane structure |
Lactobacilli and leuconostoc CAT in S. pneumoniae |
Conjunctivitis, meningitis, plague, cholera, and typhoid fever |
||
|
|
Cephalosporins (1st and 2nd generation) and cephamycins |
Cefazolin |
Cell wall biosyntesis |
Gram-positive aerobes: Staphylococcus aureus (including beta-lactamase producing strains) Staphylococcus Gram-Negative Aerobes: Escherichia coli, Proteus mirabilisepidermidis, Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus pneumoniae and other strains of streptococci |
Cellulitis, urinary tract infections, pneumonia, endocarditis, joint infection, |
||
|
|
Lincosamides |
Clindamycin |
Binds 50S ribosome, blocks peptide elongation; |
Staphylococcus aureus, Bacteroides, Fusobacterium and Prevotella, although resistance is increasing in Bacteroides fragilis. |
Dental infections and infections of the respiratory tract, skin, and soft tissue, and peritonitis |
||
|
|
Penicillins (anti-staphylococcal) |
Oxacillin |
Cell wall synthesis |
Methicillin and oxacillin resistant staphylococcus |
Respiratory or urinary tract infections |
||
|
|
Pseudomonic acids |
Mupirocin |
Inhibition of protein synthesis |
Methicillin-resistant S. aureus (MRSA) |
Superficial skin infections |
||
|
|
Riminofenazines |
Clofazimine |
guanine bases of bacterial DNA, thereby blocking the template function of the DNA and inhibiting |
Different species of Mycobacterium. |
Leprosy |
||
|
|
Steroid antibacterials |
Fusidic acid |
Protein synthesis |
Staphylococcus aureus, most coagulase-positive staphylococci, Beta-hemolytic streptococci, Corynebacterium species and most clostridium species. |
Acne vulgaris |
||
|
|
Streptogramins |
quinupristin/dalfopristin |
Protein synthesis inhibitors |
staphylococci and vancomycin-resistant Enterococcus faecium. |
Infection caused by staphylococcus and enterococcus faecium |
||
|
|
Sulfonamides, dihydrofolate reductase |
Sulfamethoxazole, trimethoprim |
Compete with p-aminobenzoic acid (PABA) preventing synthesis of folic acid |
Most of the bacteria. |
Bacterial infections (such as middle ear, urine, respiratory, and intestinal infections). |
||
|
|
Sulfones |
Dapsone |
bacterial synthesis of dihydrofolic acid, |
Leprosy, acne, dermatitis herpetiformis, and various other skin conditions |
|||
|
|
Tetracycline |
Coretetracycline |
Block tRNA binding to 30S ribosome-mRNA complex (b-static) |
Aerobic and anaerobic bacterial genera, both Gram-positive and Gram-negative, with a few exceptions, such as |
Acne, cholera, brucellosis, plague, malaria, and syphilis |
||
|
|
Important antimicrobials |
|
|
|
|
||
|
|
Aminocyclitols |
Spectinomycin |
Inhibits protein synthesis |
Bacteria |
Gonorrhoea infections, used by those who are allergic to penicillin or cephalosporins |
||
|
|
Cyclic polypeptides |
Bacitracin |
Inhibits RNA transcription |
Staphylococcus aureus –Staphylococcus epidermidis, Streptococcus pyogenes |
Skin, eye and wound infections. |
||
|
|
Nitrofurantoins |
Nitrofurantoin |
Nitrofurantoin has been shown to have good activity against: E. coli, Staphylococcus saprophyticus, Coagulase negative staphylococci, |
Bladder infections, uncomplicated urinary tract infections (UTIs |
|||
|
|
Nitroimidazoles |
Metronidazole |
Disrupts nucleic acid synthesis |
Aerobic bacteria |
Vaginal infection in women |
||
|
|
Pleuromutilins |
Retapamulin |
Protein synthesis inhibitor |
Staphylococcus aureus (methicillin-susceptible only) or Streptococcus pyogenes |
Impetigo |
||
Table 2 Following is the list of microbes important for human medicine
All the details of microbes, their antibiotic with mode of action and disease are taken from Wikipedia and corresponding drug bank.3 Different antimicrobials have different mode of action which can be diagrammatically as discussed and described in Figure 1 as:
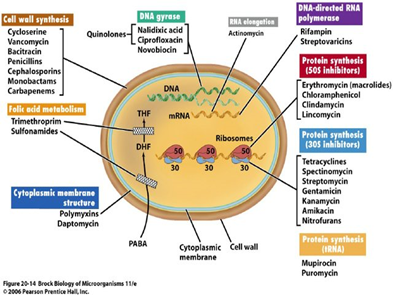
Figure 1 Mode of action of antibiotics in humans.22
Here in this paper an effort is made to classify Critically Important Antimicrobials according to the amino acid composition of responsible microbes. Because amino acids are the building blocks of proteins. And the effect of antimicrobials directly or indirectly affects proteins of microbes.
For classification of antimicrobials, machine learning (ML) techniques are employed. Because it is good in data analysis and model building. ML is a branch of artificial intelligence3–9 it makes system learn from data, identify patterns and make great decisions without human interference. As there is huge amount of variety of data computational processing are a need to understand huge data in a better way for further use. These ML computational techniques are cheaper and powerful tools to apply. Here in this paper author tries to classify and develop model for critically important antimicrobials for human medicine by support vector machines (SVM). It can be defined as a discriminative classifier means two objects or set of objects are classified by a separating hyperplane. It could be said that, as labelled training data (supervised learning) is given, the algorithm outputs an optimal hyperplane which categorizes new examples. Hence hyperplane is a line dividing a plane in two parts where in each class lay in either side in a two dimensional space.10–16
Data
In this section, preparation of training and testing dataset is described. The amino acid composition of all the protein sequences are taken from PROCOS (Protein composition server).17 It is very time consuming and accurate. Predictions of sub cellular localization of proteins are also used amino acid composition as described in 4 But due to importance of amino acids, related work was also done. It is said that the fraction of each type of amino acid type within a protein is called as amino acid composition.
equation1
After gathering all the protein sequence data which are called peptides are divided into different groups called datasets. There are three different datasets according to importance of antimicrobials.18
Datasets
Dataset 1: Critically important antimicrobials: The microbes’ protein data which is available in Uniprot database is taken. And there amino acid composition is taken by PROCOS software as input for SVM. These are called training set and are positive samples needed to be classified. For testing we took negative samples of other enzymatic group.
Dataset 2: Highly important antimicrobials: Same as dataset 1 dataset 2 is prepared.
Dataset 3: Important antimicrobials: similarly dataset 3 for important antimicrobials are also prepared.
Negative samples examples: With respect to positive samples, it requires negative interaction examples to process the positive samples accurately, as the SVM is a discriminative approach. When experimental methods do not report an interaction between two proteins, it means there positive signal does not imply a negative signal. Hence no interaction between amino acids. It is required that real negative examples are of important part for providing better results.
Feature selection with SVD: (SVD) is a method to reduce the dimensions and select the most relevant and informative features. Principal component analysis19,20 is also used for feature selection and dimensionality reduction. The higher the value of linear combination of attributes, the more important it is. For any feature corresponding eigen-value for PCA or singular value for SVD is found. Since singular value are good to choose for features. In this work SVD has lower computational cost. In SVD, the row belongs to proteins play good role in combination coefficients. In PCA the training proteins are altogether calculated the covariance between attributes. Suppose A={MO;ST} be the training dataset containing positive and negative examples, a matrix of size d*l is generated where d=p+n, it is the number of train vectors, p is the number of positive examples, n=number of negative examples, l= length of each vector. After extracting amino acid composition of different datasets, these results fed as input to Support vector machines and by performing feature selection and outlier detection. It’s important to find the hyperplane which clearly distinguish are dataset from one another with respect to their negatives. For each run of SVM the classifier is developed and their performance is measured.
Performance evaluation: The performance of our classifier was judged by 10 fold cross validation. The LIBSVM provides a parameter selection tool using the RBF kernel: cross validation via grid search. For each Dataset 1, Dataset 2, and Dataset 3 grid search is performed using c and gamma. Test set was performed for 10% of all samples and remainder samples are used for training. Generally SVM faces the problem of “over- fitting” where the system converges on the set of rules but it can be solved efficiently. The test set and train set trees are identified properly. To know the correct classification cross validation process is used. This requires for each run 10% of sample is used as test set. Different rule set up test cases are classified. It was found that which rule has the most beautiful predictive ability to improve is raised as best model evaluator. Over fitting of the data leads to the pruning.21
Machine learning algorithm for classification of antimicrobials for human medicine is implemented in this paper. All the three datasets run in LIBSVM. And best result is obtained in the form of model.
Model development
It is the final step when the data is classified as wanted. After labelling testing data and generating several classifiers. It’s final to choose which fit best classification and develop model for future use. Figure 2 shows the model for critically important antimicrobials.
According to the model development in SVM, there c,g and accuracy are calculated simultaneously and can be written in the form of Table 3 and all the required details are described later in this paper.
Dataset |
C |
G |
Accuracy |
Dataset 1 |
120 |
0.007813 |
99.8012 |
Dataset 2 |
120 |
0.0025 |
99.5 |
Dataset 3 |
120 |
0.0078 |
98.5 |
Table 3 Support vector machine results
Figure 2& Table 3 proves better that are datasets are classified accurately with great accuracy. As we focus on CIA, it was classified with 99.8012% accuracy. And also proves for similar sequences. Amino acid compositions are best suited to classify such sequences. Detail description is as follows:
Accuracy can be calculated as: =
Where tp=all the true positives in the samples
tn=all the true negatives in the sample
fp=all the samples which behave as positive
fn=those samples which behave as negative
Precision and recall, accuracy all functions are inbuilt in LIBSVM. By choosing correct c,g, software calculate all parameters and reflect the correct answer within minutes as per the volume of data. As the result obtained clearly differentiate characteristics of antimicrobials in three different groups. Any new antibiotic discovered can be grouped in above defined these categories. The correct values of c,g and accuracy of all the three datasets identified. The c and g are the two parameters for RBF kernels. It can’t be judged which is best. But the LIBSVM has the parameter selection tool which best finds the c,g, and accuracy. If good (c) is identified by the classifier then it is better prediction. The prediction accuracy indicates the performance on classifying an independent dataset. Hence it is good to know about ‘unknown” dataset. Again cross-validation is performed. In this n-fold cross-validation the training set is first divided into n-subsets of equal size. It would work sequentially by (n-1) subsets. Therefore cross validation is the percentage of data which is accurately classified. This cross validation removes the over fitting. The grid search approach is used because (a) it avoids exhaustive parameter search by approximations or heuristics, (b) Computational time is less as there is only two parameters. (c) Both c and g are independent. Hence SVM is one of the best computational methods which reduce the cost of CV and best is biological data classification.
Machine learning being an active area of research requires experts that handle data safely and understand the data as information retrieval system. Here machine learning model is developed for antimicrobials which are used in human medicine. Hence WHO initiates how to recommend critically important antimicrobials for human medicine? It’s a need to describe importance of human medicine publically. So in this paper author well tried to classify critically important antimicrobials for human medicine with great accuracy. Future treatment should be given by seeing the effect of antimicrobials. And any other microbe or antimicrobial is generated it should be grouped according to its amino acid composition based category as the machine learning model is being developed.
None.
The author declares there is no conflicts of interest.

©2018 Dubey. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.