International Journal of
eISSN: 2573-2889


Research Article Volume 3 Issue 2
Department of Biochemistry, University of Khartoum, Sudan
Correspondence: Mutaz Amin, Department of Biochemistry, Faculty of Medicine, University of Khartoum, Sudan
Received: August 07, 2017 | Published: March 1, 2018
Citation: Amin M. Candidate variants in MLC1 gene causing Megalencephalic Leukodystrophy using in silico prediction methods. Int J Mol Biol Open Access. 2018;3(2):49-52. DOI: 10.15406/ijmboa.2018.03.00049
Megalencephalic Leukodystrophy with sub cortical cysts is a type of Demyelinating Leukodystrophy caused by mutations in MLC1 gene. Various mutations in MLC1 gene have been reported worldwide but high throughput technologies aimed to discover novel variants underlying this disorder are scarce and there is a lot yet to be discovered. In silico analysis of SNPs in a gene known to cause a disease is a well effective and economic method of analyzing known variants deposited in public databases. This article aimed to analyze all SNPs in MLC1 gene in order to be used in screening programs for patients with Megalencephalic Leukodystrophy. The SNPs in MLC1 gene were retrieved from NCBI db SNP. Variants in VCF format were analyzed using Variant Effect Predictor (VEP) of the Ensemble database. The deleterious coding ns SNPs were detected by the web program SIFT, PolyPhen and Mutation Taster in addition to allele frequency and conservation score. The 3-D model of the human MLC1 protein was predicted using the CPH models 2.0 server. The resulting modeled structure with positions of mutations was viewed using Chimera 1.8 software. In MLC1 gene, 4 are nonsense, 18 are indels (frame shift mutations) and 10 potentially disrupt splicing. Six variants in MLC1 gene were predicted to be pathogenic using the same tools. The results of our study will facilitate future studies aimed to analyze the genetics of Leukodystrophy patients from different families from different populations.
Keywords: leukodystrophy, MLC1, SNPs, in silico
Leukodystrophies are group of inherited disorders caused primarily by defective myelination of the central nervous system with or without peripheral nervous system involvement.1 There are over 30 of Leukodystrophy disorders have been described with various age of onset and clinical presentation but they all share white matter signals in brain MRI.2 Individual Leukodystrophy types are rare-although they vary depending on the population- but collectively they are not uncommon.3 Megalencephalic Leukodystrophy with sub cortical cysts is a type of Leukodystrophy caused by mutations in MLC1 gene and less commonly in HEPACAM Gene.4 The disease presents with early head enlargement, motor dysfunction and occasionally epilepsy. All kinds of mutations in MLC1 gene were described.4 However, high throughput technologies aimed to discover novel variants underlying these disorders are scarce, and there is a lot to be discovered. In silico analysis of SNPs in a gene known to cause a disease is a well effective and economic way of at least screen known variants deposited in public databases. And this approach has indeed proved valuable in many situations.5‒8 Since Leukodystrophy disorders are rarely studied especially in developing countries, approaches like in silico analysis, patients can be screened for known and predicted pathogenic variants first and if none found proceed to advanced technologies like whole genome or whole exome sequencing. This article aimed to analyze all SNPs in MLC1 gene in order to be used in screening programs for patients with Megalencephalic Leukodystrophy.
The SNPs and their related protein sequences for MLC1 gene were retrieved from NCBI db SNP. Frame shift, nonsense and splicing variants were obtained from the NCBI database. The deleterious coding ns SNPs were detected by the web program SIFT and PolyPhen. Variants of both genes in VCF format were analyzed using Variant Effect Predictor (VEP) of the Ensemble database http://www.ensembl.org/Tools/VEP. Predicted pathogenic variants were filtered as follows: SIFT score <0.05, PolyPhen score >0.85 and Allele frequency <0.05. Pathogenicity of variants was verified using Mutation Taster and amino acid conservation from Alamut visual http://www.interactive-biosoftware.com/doc/alamut-visual/2.9/. Structural analysis was performed in order to evaluate and compare the stability of native and mutant structures. The 3-D model of the human MLC1 protein was predicted using the CPH models 2.0 server.9The resulting modeled structure with positions of mutations was viewed using Chimera 1.8 software.10
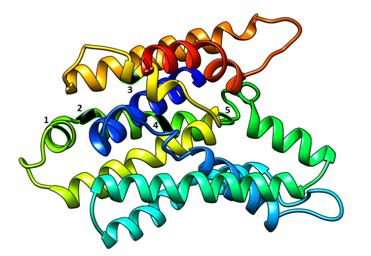
The MLC1 gene contains 1471 SNPs, majority of which are intronic (44%) and upstream variants (22%). Majority of coding variants were missense (62%) and synonymous (30%), (Figure 1). In MLC1 gene, 4 are nonsense, 18 are indels (frame shift mutations) and 10 potentially disrupt splicing, (Table 1). In MLC1 gene, 6 missense variants were predicted to be pathogenic using in silico tools, (Table 2). The result of MLC1 protein modeling revealed globular proteins with multiple helices and beta sheets (Figure 2). The position of amino acid variants mentioned above is shown in (Figure 2).
MLC1 |
||
|---|---|---|
Nonsense |
Frame shift |
Splicing |
rs992764755 |
rs4513390 |
rs992020383 |
rs992830566 |
rs4569573 |
rs992021756 |
rs992920871 |
rs4569574 |
rs992057554 |
rs993014432 |
rs4569575 |
rs992070962 |
rs4600768 |
rs992340649 |
|
rs4838816 |
rs992349844 |
|
rs4838817 |
rs992369866 |
|
rs4838819 |
rs992430530 |
|
rs4838879 |
rs992566519 |
|
rs4838880 |
rs992712599 |
|
rs4838882 |
||
rs4838883 |
||
rs4990416 |
||
rs5771140 |
||
rs5771141 |
||
rs5771142 |
||
rs5771143 |
||
rs5771144 |
||
Table 1 SNPs causing nonsense, frame shift and splicing impairment mutations in MLC1 gene
SNP |
AA* |
SIFT (score) |
Poly phen (score) |
Mutation taster |
AF** |
CS |
rs143061714 |
V260L |
Deleterious (0) |
Probably damaging (-0.987) |
Disease causing |
0.0006 |
High |
rs568289086 |
A208V |
Deleterious (0) |
Probably damaging (-0.994) |
Disease causing |
0.0002 |
High |
rs41302601 |
N218K |
Deleterious (0) |
Probably damaging (-0.997) |
Disease causing |
0.0022 |
High |
rs78644350 |
V200F |
Deleterious (0) |
Probably damaging (-0.997) |
Disease causing |
0.0002 |
High |
rs555304253 |
R193W |
Deleterious (0) |
Probably damaging (-0.997) |
Disease causing |
0.0002 |
High |
rs533294413 |
R20G |
Deleterious (0) |
Probably damaging (-0.997) |
Disease causing |
0.0004 |
High |
Table 2 Missense variants in MLC1 gene predicted to be pathogenic with SIFT (and its score) Poly Phen (and its score), Mutation Taster, allele frequency and conversation
*AA, Amino acid change; **AF, Allele frequency
In silico analysis of SNPs in disease causing genes deposited in public databases and whose clinical significance is unknown is a valuable and economic method for preliminary screening studies especially for neglected diseases like Leukodystrophy. This study aimed to which variants in MLC1 gene are likely to be pathogenic using in silico prediction methods. The most common mutation we found were indel variants causing frame shift mutations. Frame shifting mutations are known to disrupt protein synthesis and very rarely don’t impair protein function.11 Frame shifting mutations were indeed found in many studies underlying these forms of Leukodystrophy.12‒15 Splicing impairing variants and non-sense variants were also found in our study and from pathogenesis point of view they are even stronger as culprits than either frame shift or missense damaging variants especially for disease which loss of function is their known mechanism16. Nonsense variants were found also in other studies17‒19 and so are splicing defects.15,20 Missense variants causing MLC1were previously reported in many studies.21 Judging from the in silico prediction methods which include conservation, allele frequency and tolerance of amino acid changes, our variants reported in our study are very likely to be pathogenic, but they are all novel and their association with these types of leukodystrophies await to be confirmed. These variants were deposited in NCBI public database with unknown clinical significance because all individuals sampled for genotyping were in heterozygous state and these diseases are recessive. The results of our study will facilitate future studies aimed to analyze the genetics of Leukodystrophy patients from different families from different populations. By implementing in silico predicted pathogenic variants in disease panels, this will perhaps lower the need to do exome sequencing and hence the cost of the study. The results of in silico predictive studies will also accelerate variants annotation keeping in pace with advancing international genome and exam studies.
None.
The authors declare that they have no conflict of interest.

©2018 Amin. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.