International Journal of
eISSN: 2573-2889


Pakistan is a country with over 210 million people largely practicing cousin marriages (consanguinity). More than 60% of Pakistanis are born to consanguineous marriages; the risk of autosomal recessive disorders is 18 times higher in consanguineous marriages, compared to unrelated couples. This population is at a higher risk of having children with genetic disorders due to inbreeding and fast growing population. Inherited disorders, in general are incurable; however, early genetic testing and screening may help reducing the affected births. Mental retardation (MR) or Intellectual disabilities (ID) is a complex and heterogenic term occupying several clinical directions. Mental retardation (MR) or Intellectual disabilities (ID) is a complex and heterogenic term occupying several clinical directions. Defining features of MR include sub average general intellectual functioning with deficiency in at least two of self-survival skills that appeared during childhood i.e. before the age of 18 years.1 Both genetic as well as environmental causes play their role in causing MR and disturb the development and functioning of central nervous system prenatally, prenatally or postnatal.2 Mental retardation is being responsible to affect about 1-3% of the general population.3 Phenotypically MR can be syndrome where MR is accompanied additional malformation/s, dimorphic features, or neurological abnormalities, and non-syndrome (NS-MR), which are specified by MR without any additional dimorphic feature. High prevalence in males than females supports the idea of involvement of many x-linked genes in NS-MR.4 The total number of genes involved in NS-MR may exceed more than a thousand which are not surprising since about half of the estimated 25,000 human genes are expressed in the brain.5 NS-MR is one of the serious health problem with neuro developmental abnormality, having grim impact not only on the affected individuals and their families but also agitate the health care system and society. Knowledge and understanding about the genes and loci involved in nonsyndromic autosomal recessive mental retardation is not satisfactory yet.
Large consanguineous Pakistani family with NS-ARMR was ascertained from Punjab region of Pakistan. All patients in the family were having phenotypes of low IQ and learning disability without any neurological, physical and biochemical abnormalities. Family was subjected to autozygosity mapping for all know NS-autosomal recessive mental retardation (ARMR) genes using STS (Sequence tagged sites) markers followed by autozygosity mapping using microarray single-nucleotide polymorphism (SNP) analysis. Several candidates’ homozygous regions including 6-Mb telomere region on 11p15 were obtained for the nuclear central loop in the five generation family. Except 6-Mb telomere region on 11p15, all other linkage regions were debarred by calculation of logarithm of odds (LOD) for the SNP microarray data (Figure 1A) (Figure 1B). A maximum LOD score of Z=3.31 was calculated for the mapped region (Figure 1C), showing potential likeliness of the region to be inherited together. These results recommend a novel genetic locus, MRT17, for NS-ARMR.4 The reported locus called MRT17 contains different genes including PGAP2; encodes for protein named Post Glycosyl Phosphatidyl Inositol (Post-GPI) attachment to proteins factor 2, and having interaction with genes involved in mental processes. Based on this surface information, PGAP2 was assumed to be mutated; hence resulting in altered protein, causing cognitive impairment in patients. Another family showing overlapping to MRT17 linkage region with same phenotypes was also observed.6 Both families harbour one homozygous mutation in PGAP2 gene encoding a protein implicated in the remodelling of the GPI anchor in the Golgi apparatus. PGAP2-deficient Chinese hamster ovary (CHO) cell lines were used for functional analyses, showing reduced activity of the respective altered proteins; hence confirming pathogenicity. Autozygosity mapping and ultra-deep sequencing followed by stringent filtering were performed, showing two homozygous alterations in PGAP2, p.Tyr99Cys and p.Arg177Pro, in seven patients with nonspecific AR-MR from both consanguineous families. Rescue experiments was performed with the altered proteins in PGAP2-deficient Chinese hamster ovary cell lines and results showed reduced expression of DAF and CD59 (cell-surface GPI-anchored proteins) than of the wild-type protein, validating the pathogenicity of the observed alterations. Furthermore, full rescue was observed by using strong promoters before the mutant cDNA’s, signifying a hypo orphic outcome of the mutations.6 Alterations in the Golgi-located part of the GPI-anchor-biosynthesis pathway were reported and broaden the phenotypic range of the GPI-anchor deficits to isolated intellectual disability with raised Alkaline Phosphatase (ALP). All such observations are strengthening the concept of involvement of group of GPI-anchor deficits in disease causing within the family as provided by Brunner and van Driel,7 and it was hypothesized that harshness of the phenotype of a GPI-anchor deficits is reliant on the position of the altered protein in the synthesis network. Transcript variant 12 cDNAs were sub cloned; encoding wild-type and altered iso form 8, using mammalian expression vector with three different promoters: strong SR promoter, medium-strength TK prom and weak TATA promoter. These findings revealed that p.Tyr99Cys and p.Arg177Pro altered PGAP2 proteins have considerably down particular role in the pathway. PGAP2 encodes a protein in the biosynthesis network of the GPI anchor with probable structure of five transmembrane alpha-helix domains and is positioned in the Golgi apparatus8 (Figure 2). Alteration of strongly hydrophobic nature Arginine to non polar proline (p.Arg177Pro) and hydrophobic nature tyrosine to the hydrophilic nature cysteine (p.Tyr99Cys) in the Golgi lumen imply alteration in the secondary and tertiary structures of the protein and might cause disturbance in protein folding. These alterations can also conceptualize disturbance in the PGAP2 interaction with a so far unidentified putative acyltransferase.
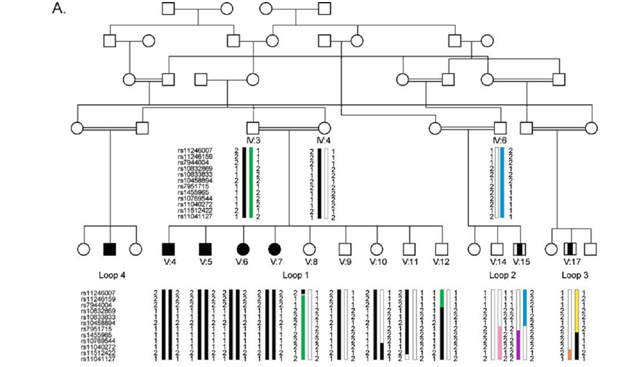
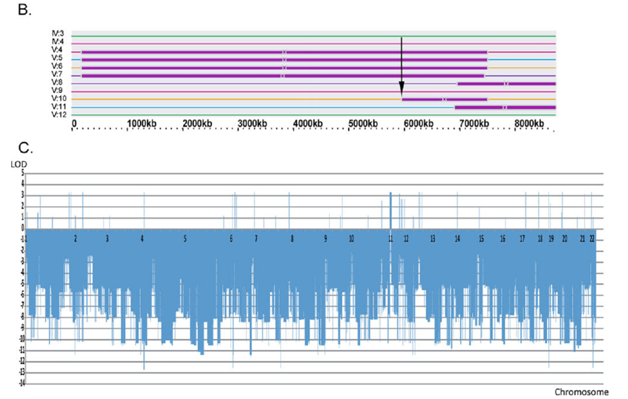
Figure 1(A) Large consanguineous Pakistani family with AR-NSMR, showing haplotypes for 6 Mb telomeric linkage regions on 11p15.
(B) Graphic presentation of the homozygous telomeric linkage region on 11p15 with recombination breakpoint found for V:10 shown by an arrow.
(C) The LOD scores calculation for different chromosomal regions of the entire genome using the programs LIP-SNP6 and LIPED.
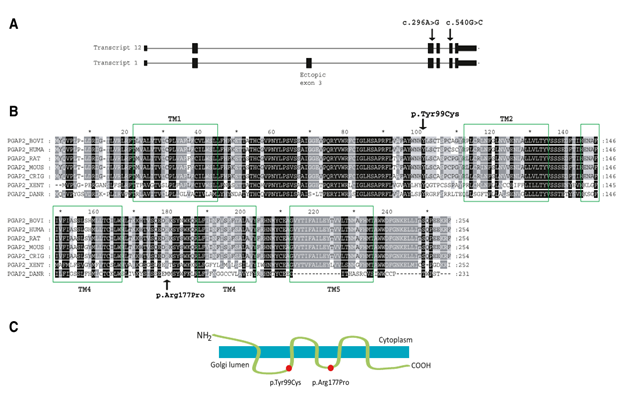
Figure 2 (A) PGAP2 structure showing Exons-Introns combination. Arrows indicate mutations. (B) Sequence of human PGAP2 and its alignment to other organisms. (C) PGAP2 encodes a transmembrane protein, and the p.Tyr99Cys and p.Arg177Pro changes are positioned in the Golgi lumen.
The GPI-anchor-biosynthesis network includes more than involves over 20 different proteins. On completion of synthesis, GPI anchors are subsequently shifted to the proteins in the endoplasmic reticulum (ER) and subjected to additional modification by the elimination of acylphosphate and ethanolaminephosphate chains. With the help of Golgi apparatus, GPI-APs (Anchor proteins) are further transfererd from the ER to the plasma membrane. Fatty acid remodelling of the GPI anchor comes about in the Golgi apparatus; an unsaturated fatty acid is removed by PGAP3, and a saturated fatty acid is transported back, much probably by non catalytic PGAP2 protein. Most likely, an uncharacterized acyltransferase can play its role in back transportation provided that PGAP2 itself does not resemble other acyltransferases.9,10 The GPI-anchor deficits demonstrate a subtype of congenital diseases of glycosylation.11 Six genes with germline mutations; encoding proteins in the ER-located part of the GPI-anchor-biosynthesis network are reported: those in PIGA (MIM 311770), PIGL (MIM 605947), PIGM (MIM 610273), PIGV (MIM 610274), PIGN (MIM 606097), and PIGO (MIM 614730).6 Phenotypes of PIGA, PIGL, and PIGN mutations consequent in protein alteration are coupled with abnormalities and in some cases are even fatal. Mutations in PIGV and PIGO cause further disturbance in the pathway and lead to intellectual disability, hyper phosphatase, epilepsy, and minor dysmorphisms.6 All mutations are recessive with residual protein activity, and it looks like that absolute deficit in GPI-anchor biosynthesis is mismatched with life.12 A single known mutation in a regulatory element of PIGM causes specific phenotype of portal and hepatic vein thrombosis and epilepsy.13 All these events and discussion conclude that hypomorphic mutations in PGAP2 cause altered protein product; leading to severe intellectual disability with raised ALP and no apparent malformations. This signifies a blemish in the GPI-anchor-biosynthesis step occurring in the Golgi and engages remodelling of the anchors, and it spreads out the phenotypic range of GPI-anchor deficits to isolated intellectual disability.
None
Author declares there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.