eISSN: 2471-0016


Research Article Volume 6 Issue 2
1Institute of Experimental Medicine, Central University of Venezuela, Venezuela
1Institute of Experimental Medicine, Central University of Venezuela, Venezuela
2Anatomopathological Institute Jos
2Anatomopathological Institute Jose A. O Daly Central University of Venezuela, Venezuela
3University Hospital of Caracas, Central University of Venezuela, Venezuela
3University Hospital of Caracas, Central University of Venezuela, Venezuela
44nstitute of Tropical Medicine, Central University of Venezuela, Venezuela
44nstitute of Tropical Medicine, Central University of Venezuela, Venezuela
Correspondence: Rossi S Marcello Salvatore, Section of Immunohistochemistry and Electron Microscopy, Jose A. O Daly Anatomopathological Institute, Faculty of Medicine, Central University of Venezuela, Calle Alejandro Casona 7, 6D Floor, Madrid (28035), Spain, Tel +3491 1265 512
Received: January 01, 1971 | Published: March 7, 2018
Citation: Correa de Adjounian MF, Rossi S MS, Chalbaud A,et al. Importance of using several molecular targets for the appropriated identification of Mycobacterium marinum. Int Clin Pathol J. 2018;6(2):67–74. DOI: 10.15406/icpjl.2018.06.00159
M. marinum is a non-tuberculous mycobacterium capable of producing human infection with variable incidence, often under diagnosed due to the difficulties of isolation and culture. There is a description of a M. marinum infection in which the molecular identification required the use of several polymorphic sequences. From ulcerated papules and purulent discharge patient lesion histopathology, bacteriological identification by biochemical tests and molecular identification using PCR-restriction fragment length polymorphism analysis (PRA) of 3 polymorphic sequences with two regions of hsp65 gene and an
spacer of 16S-23S rDNA was performed.Histopathological studies showed granulomatous lesions with inflammatory cell infiltrate. The isolation and bacteriological identification showed the causal agent to be M. marinum. However, the initial genotypic identification, using mostly polymorphic sequence of the hps65 gene, did not discriminate from M. marinum and M. malmöense type I, requiring the use of other PRA to confirm the diagnosis of M. marinum. The joint application of bacteriological and molecular techniques established the diagnosis. We suggest that there is necessary to include more than one molecular target to achieve the identification of these mycobacteria species.
Keywords: Non-tuberculous Mycobacteria (NTM), Mycobacterium marinum, hsp65, 16S-23S rRNA, PCR-restriction fragment length polymorphism analysis (PRA)
PRA, PCR-restriction fragment length polymorphism analysis; hsp65, gene for the 65 kda heat shock proteins; KDa, kilodalton, rDNA, ribosomal deoxyribonucleic acid; NTM, non-tuberculous mycobacteria; MTC, mycobacteria of tuberculous complex; USA, United States of America; PCR, polymerase chain reaction; rRNA, ribosomal ribonucleic acid; ZN, ziehl-neelsen stain; LJ, Löwestein-Jensen medium; HE, Haematoxylin-Eosin stain; TE buffer, Tris-EDTA buffer; Ejk, Euclidean distance; LCR, ligase chain reaction; LAMP, Loop-Mediated Isothermal amplification; bp, base pairs
Non-tuberculous mycobacteria (NTM) are environmental free-living microorganisms very common in soils and waters of terrestrial and aquatic ecosystems or being part of the commensal or pathogenic bacterial flora from wildlife, domestic and marine animals.1 Although interactions of human being with the NTM are very common, they infrequently cause symptomatic infections in immunocompetent individuals. For this reason, infections caused by these mycobacteria have received less attention than those produced by mycobacteria from tuberculosis complex (MTC) such as Mycobacterium tuberculosis, M. bovis, M. africanum, M. caprae and M. microti, among others. In addition, infections caused by NTM are not of mandatory declaration to the public health agencies of some countries. Only recently NTM have received some clinical attention due to the complications they cause to immunosuppressed individuals, as in the case of patients infected with the human immunodeficiency virus (HIV) or under treatment with steroids or antineoplastic agents. The number of NTM species of clinical importance increases every day, and species such as M. avium, M. intracellulare, M. scrofulaceum, M. fortuitum, M. chelonae, M. malmöense, M. kansassi, M. gordonae and M. marinum among others, have been described as causative agents of diverse mycobacteriosis, characterized by a wide range of focal or systemic clinical manifestations that have being become in a medical challenge in terms of diagnosis and treatment.2–4
The identification and differentiation of these mycobacteria at species-level is mainly carried out based on phenotypic methods where bacteriological techniques are included. Despite these methods require the investment of considerable man-hours, multiple tests and moderately expensive reagents; they have as main advantage the partial differentiation of MTC and NTM with relative certainty.5 After bacteriological methods were implanted the chromatographic methods have evolved in order to improve mycobacterial identification but despite these techniques are tools of high precision, they require very specialized laboratories and very expensive equipment.6 Although molecular methods require also specialized laboratories and resources, they are more popular and accessible, existing an increasing trend to use them in laboratories but without displacing conventional methods of culture and biochemical identification as the first approach to the identification and amplification of mycobacteria populations.7 Furthermore, molecular tests are fundamental since the selection of drugs and treatment schemes against mycobacterial diseases are based on the genetic identification of the mycobacterial species involved.8
As a mycobacteria grouped into NTM, M. marinum is considered as saprophyte specie, capable not only to infect about 150 species of freshwater and saltwater fish, but also to produce infections in the soft tissues of human being.9 Despite it was isolated for the very first time in 1926; the first reports of human infections with this mycobacterium were associated with recreational activities in pools or spas with poor hygiene measures.2 In this regard, the isolation of this specie has been described in a 4.5% of the floor of pool showers analysed.10
In addition, there is increasing evidence that the infections could occur among people engaged in aquaculture and aquarium-keeping activities,11 as well as in sport and commercial fishing, and fish marketing.12 In most of cases infection is established when penetrating traumas occur, especially in the upper extremities, either by fish bites or by injuries produced by crustacean chelae, spines or fish bones, as well as due injuries caused by fishing equipment such as hooks and contaminated harpoons, or the handling of exotic pets.13 The incidence of M. marinum infections is quite variable. In this regard, studies conducted in North Carolina (USA) have estimated the incidence of these infections as 27 cases per 100,000 adult inhabitants.9 A multi-centre study conducted in Spain, revealed that the incidence of human infections by M. marinum is 0.5 cases per 100 cases of NTM infections.14 The very low incidence of infections by M. marinum could be related to the fact that this mycobacterial species is often underdiagnosed due to the indolent nature of its clinical presentation, the difficulties of its isolation and its culture as well as the unawareness of its clinical profile by many physicians and infectologists. Based on this, the recognition of M. marinum requires a high index of suspicion by the clinician or infectologist and a careful study of the clinical history in order to reveal the patient's exposure to marine products, fishing or other aquatic activity.15,16 The identification of M. marinum by bacteriological methods is achieved through biochemical, physiological and pigment production tests. In this regard, it is a photochromogenic mycobacterium (Runyon Group 1), of rapid growth (7-14 days at 32 °C), which forms smooth and flattened colonies with irregular edges that give weak positive results to the catalase production at 25°C, strong positive result to the production of urease and negative result to the production of niacin and nitrates. Despite it has been described that these tests allow differentiating M. marinum from M. kansasii and other NTM of rapid growth, 2 in some cases differentiation of M. marinum from other mycobacterial species of the Runyon Group 1, not only is a very hardworking procedure but also could give uncertain results. For these reasons, molecular techniques such as the polymerase chain reaction (PCR) combined with the restriction analysis of the corresponding amplification products (PRA), has proved to be a useful tool in the mycobacterial identification. For these purposes several molecular targets have been used, such as the hsp65 gene17–19 and the ribosomal intergenic,20 among others, for the identification of M. marinum and its differentiation from other NTM.
In this work we describe a case of mycobacteriosis caused by M. marinum misdiagnosed as M. malmöense according to the restriction pattern obtained when using a unique sequence of hsp65 gene. Results are discussed on the basis of the importance of using more than one gene sequence of hsp65 gene and the intergenic 16S-23S non-transcribed sequence of rRNA for an accurate identification of this species.
Patient
A 43-year-old male patient from the town of Río Chico (Miranda State, Venezuela), attended the Dermatology Service of the University Hospital of Caracas at the Central University of Venezuela on September 2001 due to the presence of hyperpigmented papules, nodules and plaques disseminated in the upper right arm (Figure 1A). Some of these lesions showed ulcers and spread to the forearm, hand and fingers (Figure 1B). Samples of purulent secretion were taken from ulcerated papules to be used in bacteriological and molecular studies. In the anamnesis the patient reported that the lesions began to develop in June of the same year and that he is engaged on fishing. Among the clinical records the most important were Cushing's disease, alcoholism and left supracondylar amputation.
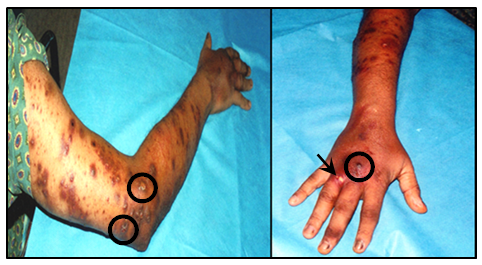
Figure 1 Skin lesions of the patient showing (A) Multiple violaceous and dark papules with a sporotrichoid pattern of distribution at arm level and linear at forearm level, following the lymphatic drainage pattern of the limb. Notice that some lesions were ulcerated (○). (B) Lesions on the dorsal side of the hand and fingers, with the presence of a nodule at the level of middle finger phalange (○) and an ulcerated lesion at level of the phalange of the ring finger (→).
Bacteriological studies
Samples of the purulent secretion were referred to the Laboratory of Bacteriology of the Institute of Tropical Medicine (Central University of Venezuela), where aliquots were used in order to prepare smears stained by the Ziehl-Neelsen (ZN) technique and to inoculate Löwestein-Jensen medium (LJ). The cultures with mycobacterial growth were used for biochemical identification (growth at 32 °C, growth in sodium chloride, production of pigments, arylsulfatase test, Tween hydrolysis, and nitrate reduction). Moreover a sample of them was referred to the Reference National Centre for Tuberculosis at El Algodonal Hospital.
Histopathological studies
Skin biopsies obtained from the lesions were fixed with 10% buffered formalin and then processed by conventional histological technique. Histological sections 4 μm-thicks were stained by the Haematoxylin-Eosin (H-E) technique and permanent preparations were analysed under microscope. The evaluation of the biopsies showed the presence of histiocytic granulomatous lesions with mononuclear inflammatory cells infiltration at the level of the dermis (Figure 2A). The ulcerated lesions from which the sample was obtained were characterized by the presence of abundant mononuclear inflammatory cells (lymphocytes, plasma cells), oedema and the presence of granulomas (Figure 2B).
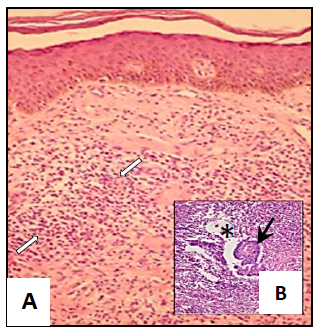
Figure 2 Photomicrographs of studies from skin biopsies showing (A) Inflammatory processes of dermis characterized by leucocytes infiltration (⇨⇦) (Amplification: 10 X) and (B) Groups of giant cells (Langhans cells) forming granulomas (→) in oedematous spaces (*) with acidophilic necrotic remains surrounded by inflammatory cells (Amplification: 40X).
Molecular studies
Molecular identification of mycobacteria present in the lesions was made with purified DNA from mycobacteria isolated in LJ medium. The DNA was extracted according to the method described by Kolk et al.21 Briefly, mycobacterial colonies were suspended in TE buffer, inactivated at 100°C for 30 minutes and bacterial walls and cellular proteins were digested with lysozyme and proteinase K respectively. DNA was then extracted with a mixture of chloroform: isoamyl alcohol (24:1) and precipitated with cold isopropanol after centrifugation at 13,000xg. The genomic DNA obtained was dried at ambient temperature and suspended in TE.
Polymerase chain reaction (PCR) and restriction analysis of amplification products (PRA)
Mycobacterial genomic DNA was used as a target for PCR amplification of two different polymorphic sequences of the hsp65 gene17,18 and one corresponding to a segment of the ribosomal intergenic non-transcribed spacer (16S-23S rRNA) [20]. The 439 bp amplification product of the hsp65 gene (PCR1) was restricted with the enzymes BstEII and HaeIII, while a fragment of 295 bp corresponding to the same gene and amplified during PCR2, was restricted with the enzymes CfoI and Sau96I. The restriction analysis of the amplicons obtained from ribosomal intergenic spacer (PCR3) was performed with the enzymes CfoI, HaeIII and TaqI. In all cases, the restriction fragments were separated by electrophoresis on 4% NuSieve agarose gels (3:1) and visualized after staining with ethidium bromide.
The size of the restriction products was estimated using the 25 and 100 bp ladders and the ɸX174 DNA restricted by HindIII (Promega®) as electrophoretic mobility patterns. The estimation of restriction fragments size was achieved using the software Q.4.1 (BioRad®). Once the size of the fragments was determined, data obtained for the 439 bp sequence of the hsp65 gene was entered into the PRA-site website of the Pasteur Institute of France,22 where size of the fragments obtained for the clinical samples were compared with those obtained by the same technique for reference strains of species of genus Mycobacterium. The degree of adjustment between the restriction patterns obtained and those showed by the reference strains, was determined through a similarity analysis based on the calculation of the Euclidean distance as:
According to this analysis, while lowest the value of the Ejk calculated between two mycobacteria is, greater is the degree of similarity between them.
Results
Histopathological studies
Studies of skin biopsies showed the occurrence of an inflammatory process of the dermis characterized by areas of mixed infiltration (Figure 2A) with abundant lymphocytes, plasma cells and histiocytes. Additionally, the presence of groups of giant cells (Langhans cells) could be observed forming granulomas (Figure 2B) surrounded by inflammatory cells. These structures were characterized by oedematous spaces, acidophilic necrotic remains and a centre containing alcohol-acid resistant bacilli (results not showed) (Figure 2B).
Bacteriological studies
Table 1 summarizes the results of the biochemical tests carried out for the bacteriological identification of the isolated mycobacteria from the purulent secretion. As it can be seen, the mycobacteria were characterized by a rapid growth (7-14 days) with an optimal growth temperature of 32 °C, and the capability to produce pigments in the presence of light. Furthermore, they present arylsulfatase activity, hydrolyse Tween 80, are unable to grow in medium containing sodium chloride and reduce nitrates (Table 1).
Tests |
Cultures from Patient´s Purulent Secretion |
M. marinum |
M. kansasii |
Speed of growth in LJ medium* |
7-14 days |
7-14 days |
7-14 days |
Optimum growth temperature |
32 °C |
32 °C |
37 °C |
Growth in presence of NaCl |
- |
- |
¿? |
Pigments production** |
+ |
+ |
+ |
Arilsulphatase activity |
+ |
+ |
± |
Tween 80 hydrolysis |
- |
- |
+ |
Nitrate reduction after 14 days |
- |
- |
+ |
Niacin reduction |
ND |
- |
- |
Catalase activity heat stable |
ND |
- |
+ |
Table 1 Summary of the biochemical and physiological performed for the differentiation of M. marinum from M. kansasii. Tests applied to the cultures of the mycobacteria present in the purulent secretion are shown in bold
*Defined as the time that elapses between the inoculation of the medium and the appearance of the first colonies
**Defined as the ability of mycobacterial colonies to produce pigments from the carotenoid family when growth is allowed in the presence of light.
Molecular studies
The PCR products of the two sequences of the hsp65 gene, amplified from genomic DNA of M. tuberculosis (H37Rv) and from the mycobacteria isolated from patient´s purulent secretion, are represented by fragments of 439 bp (PCR1) and 294 bp (PCR2) (Figure 3A) (Figure 3B). The products of amplification of the intergenic spacer 16S-23S of rDNA (PCR3) are shown in Figure 3C. Figure 4 shows the restriction fragments of the amplification products of PCR1 (A), PCR2 (B) and PCR3 (C) obtained for each of the samples analysed (Figure 3). As it can be seen in Figure 4A, after restriction of the 439 bp fragments obtained during PCR1 with the BstEII enzyme, two 230 and 203 bp bands were revealed in the sample obtained from the patient. Restriction of the 439 pb product with HaeIII enzyme generated 3 bands of 141, 107 and 82 bp and 2 bands of <50 bp that are not considered in the analysis. Table 2 shows the final result of the restriction analysis of the 439 bp fragments from hsp65 gene. Despite the tight similarity among the polymorphic patterns obtained for the sample and M. marinum type I (Ejk = 13), identification based on this amplicon did not show a clear differentiation among M. marinum and M. malmöense as deduced by the similarity existing among the polymorphic patterns obtained for the sample in relation and M. malmöense type 2 (Ejk = 13). The similarity of the sample polymorphic pattern and that described for M. kansassi type I (Ejk = 17) was also high, however Euclidean distances obtained by comparison of the pattern from the mycobacteria present in the purulent sample with M. branderi type I (Ejk = 25) and M. peregrinum (Ejk = 26) was low enough to allow to differentiate them. PRA performed with the 294 bp amplification product from hsp65 gene (PCR2), determined two bands of 225 and 70 bp after restriction with the enzyme CfoI and two of 175 and 50 bp after incubation with Sau96I (Figure 4B). Enzymes HaeIII, CfoI and TaqI, used in the PRA for PCR3 product, did not produce restriction of the corresponding amplicon, determining a unique and unrestricted 200 bp band (Figure 4C). In the case of these molecular targets, the restriction polymorphic fragment patterns were found to be compatible with M. marinum as it has been described by Telenti et al.,17 Devallois et al.18 and Park et al.20 (Figure 4) (Table 2).
Size of Fragments (Bp) Obtained With |
||||
ID |
Species/Sample |
BstEII |
HaeIII |
Euclidean distances (Ejk) |
13 |
M. marinum type1 |
240/210/0 |
145/105/80 |
13 |
104 |
M. malmöense type2 |
240/210/0 |
145/105/80 |
13 |
17 |
M. kansasii type1 |
240/210/0 |
130/105/80 |
17 |
60 |
M. branderi type1 |
240/210/0 |
140/120/100 |
25 |
66 |
M. peregrinum type2 |
240/210/0 |
145/105/60 |
26 |
WN |
Sample (295M) |
230/203/0 |
141/107/82 |
0 |
Table 2 Restriction analysis on the 439 bp fragment amplified from the hsp65 gene during PCR1. The size (bp) of the restriction fragments generated by the enzymes BstEII and HaeIII on a sample from mycobacteria cultures from patients purulent sample and reference strains of M. marinum, M. malmöense, M. kansasii, M. branderi and M. peregrinum, as well as the Euclidean distances calculated by algorithm from Pasteur Institute of France are shown
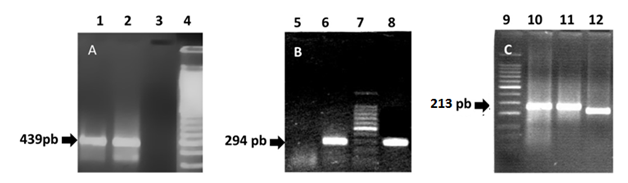
Figure 3 1% Agarose gels stained with ethidium bromide showing (A) the products of PCR1 for the amplification of the 439 bp fragment from hsp65 gene, (B) PCR2 for the amplification of a 294 bp fragment from the same gene and (C) 3% Agarose gel stained with ethidium bromide showing a fragment amplified from the intergenic spacer between the 16S and 23S genes of the rRNA. Lanes 1, 8 and 12 (DNA of mycobacteria isolated from the patient), Lanes 2, 6 and 10 (DNA from M. tuberculosis H37Rv, Positive control), Lanes 3 and 5 (Negative controls) and Lanes 4, 7 (100 bp DNA ladder), Lane 9 (50 bp DNA ladder) and Lane 11 (DNA of a mtp40+ and IS6110+ mycobacterial isolated).
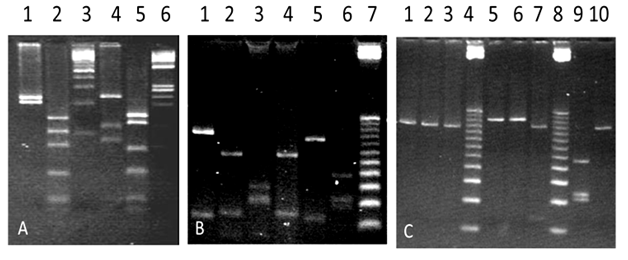
Figure 4 Gels of 4% NuSieve agarose stained with ethidium bromide showing the restriction fragments of: (A) 439 bp from hsp65 gene (PCR1) with BstEII (Lanes 1 and 4) and HaeIII (Lane 2 and 5). Lanes 1 and 2 (DNA of mycobacteria isolated from the patient), Lane 3 (100 bp DNA ladder), Lanes 4 and 5 (DNA from M. tuberculosis H37Rv, Positive control), Lane 6 (DNA ladder from ɸX174 restricted with HaeIII), (B) 294 bp amplified from hsp65 gene using CfoI (Lanes 1, 3 and 5) and Sau96I (Lanes 2, 4 and 6). Lanes 1 and 2 (DNA of mycobacteria isolated from the patient), Lanes 3 and 4 (DNA from M. simiae), Lanes 5 and 6 (DNA from M. tuberculosis H37Rv, Positive control) and Lane 7 (25 bp DNA ladder) and (C) the amplification product for the 16S-23S intergenic spacer gene of the rRNA; with HaeIII (Lanes 1, 5 and 9), CfoI (Lanes 2, 6 and 10) and TaqI (Lanes 3 and 7). Lanes 1, 2 and 3 (DNA from the mycobacteria isolated from the patient), Lane 4 (25 bp DNA ladder, Lanes 5, 6 and 7 (DNA of a mycobacterial isolated mtp40+ and IS6110+), Lane 8 (25 bp DNA ladder), Lanes 9 and 10 (M. tuberculosis DNA H37Rv, Positive Control).
At present the diagnosis and taxonomic identification of the different species grouped within the genus Mycobacterium is carried out according to phenotypic methods (i.e. microscopy, culture, bacteriological tests, Mantoux or tuberculin test, mycolic acid profile and immunohistochemistry) as well as genotypic methods such as hybridization and amplification of specific mycobacterial gene sequences by LCR, PCR, LAMP, PCR-restriction fragment length polymorphism (PRA). Results of bacteriological tests performed on mycobacteria isolated from patient´s purulent secretion are compatible with the identification as M. marinum, according to what is described in the culture manuals and biochemical identification of mycobacteria.23 Despite M. marinum and M. kansasii are fast-growing and photochromogenic mycobacteria, M. kansasii grows best at 37°C, does not hydrolyse Tween 80 and reduces nitrates. In this sense, the absence of nitrate reduction2 allowed to identify the mycobacteria from clinical samples as M. marinum with a probability of 95% according to the recommendations of Kent & Kubica.23
Although bacteriological tests have been considered as "gold standard" due their high sensitivity, specificity and predictive value, in some cases, especially in those in which mycobacteria show similar physiological and biochemical features, mycobacteriology have required help from other phenotypic or molecular techniques for the confirmation of its results. According to these facts, and to cite some examples, the differential identification of M. kansasii and M. marinum can be difficult because 95% and 80% of the isolated from each species give negative results to the tests of nitrates reduction and semi-quantitative catalase as well as to those oriented to assay drugs sensitivity.24 On the other hand M. simiae (a mycobacterium included into the tuberculous group) can be considered similar to M. tuberculosis because more than 85% of strains give positive results to the niacin test on mature colonies growing in Löwestein-Jensen. Moreover, colonies of both species are capable to show a weak yellow pigmentation that only intensifies after an extended incubation. Differentiation of M. fortuitum and M. chelonae by bacteriological tests can be also difficult, due colonies of M. chelonae can be indistinguishable from those of M. fortuitum in the physiological tests of 3-day arylsulfatase and growth on McConkey agar. Among the genotypic methods, techniques based on the amplification of mycobacterial gene sequences by PCR, or those coupled to restriction of PCR amplicons (PRA) or hybridization of PCR products with mycobacterial genomic DNA25 have been widely used in the identification. Techniques based on sequencing of polymorphic regions were also used more recently.26,27 In this regard, different molecular targets from Mycobacterium genome have been exploited, such as the gene coding for the 32 KDa protein,28 the rpo gene,29 the gene of 16s subunit of ribosomal RNA,30,31 the hsp65 gene that encodes for the 65 KDa heat shock protein,17,18,32–34 the IS6110 insertion sequences35,36 exclusive of the tuberculous complex and the mtp40 sequence37,38 apparently unique in M. tuberculosis. In our laboratory NTM identification based on molecular targets such as genic sequences of hsp65 gene and the 16S-23S non-transcribed intergenic ribosomal spacer, have been widely explored and compared with the bacteriological techniques. According to results of our studies, correlation between the identification of NTM with these molecular targets and bacteriology using clinical samples from different origins (bronchial lavage, biopsies, sputum, urine, pus of abscesses, blood, bone marrow and other body fluids), has been estimated in 93%.39 On the other hand, these molecular targets have been used successfully for the identification of mesotherapy-associated mycobacterial and nocardial infection40 as well as for confirmation of mycobacterial aetiology in autopsy cases.41
The case herein presented is an example of how genotypic techniques confirm the results obtained by techniques of phenotypic identification such as bacteriological tests. Regarding to Mycobacteria species associated with the case presented, M. marinum is a mycobacteria initially designated as M. balnei, for having been discovered in old pools and beaches with little sanitation and hygiene. In addition is the causative agent of a mycobacteriosis associated with infections of the skin, lymph nodes and non-respiratory samples with periods of incubation that vary between 4 to 8 weeks.42 From the clinical point of view, it is grouped with pathogens such as M. ulcerans, M. chelonae and M. haemophilium, among others, due its occurrence in cutaneous infections of anatomical sites with low body temperatures, as well as in association with HIV infection.2 In Venezuela the prevalence and incidence of M. marinum infections in human populations, as well as in crustaceans, molluscs and fish for domestic consumption is unknown. However in a previous study conducted by our group on 256 clinical samples from 188 patients referred to the Institute of Tropical Medicine (Central University of Venezuela), it was obtained that the joint application of bacteriological and molecular tests on 41 mycobacterial isolations allowed to identify a 37.2% (16 isolations) as NTM from which a 25% (4 out 41) were identified as M. marinum, 37.5% (6 out 16) as M. chelonae, 18.75% (3 out 16) as M. fortuitum and 18.75% (3 out 16) as undetermined mycobacteria. On the other hand, the extensive coastal geography and water basins as well as the presence of susceptible hosts, animal reservoirs and economic activities based on artisanal fishing, aquaculture and aquatic sports activities, highlight the potential risk of human communities to infection with this mycobacterium.
The estimation of the incubation period of infection herein presented was not possible; however an aspect of infection that agrees with that reported in the literature is the anatomical location and clinical evolution of the lesions. According to Ryan & Bryant43 lesions caused by M. marinum are predominantly located in the skin of the upper limbs, being characterized by their nodular, warty, bluish and ulcerated aspect. Additionally, they showed a sporotrichoid pattern of dissemination, proliferating along the path of lymphatic drainage of the affected limb. The histopathological studies performed on skin biopsies from the affected patient, allowed to verify that lesions presented different degrees of evolution, being able to recognize early lesions characterized by a mixed inflammatory response with suppuration in some cases, and old lesions that presented as well-organized granulomas. Despite the skin is the body organ most commonly involved in infection, this mycobacterium has been also identified as responsible for the arthritis and tenosynovitis of hands and wrists. Many of these infections are usually aggressive, causing permanent damage of joints, extensive osteomyelitis and amputation of the involved limb in severe cases, pathological processes that could explain the supracondylar amputation of the patient´s left arm that is recorded in patients anamnesis.
In immunocompetent patients, the lesions have a tendency to heal spontaneously after several months, although in some cases they may persist for many years. Disseminated lesions, such as those presented in this case, are characteristic of immunosuppressed patients, a condition that is supported by the clinical record of Cushing's disease and the patient's alcoholism. Most M. marinum infections are associated with penetrating traumas caused by the fish skeleton, crustacean exoskeleton and the tools used for fishing. Although the distribution of human infections are independent of sex, infections of men are strong correlated with the occupational exposure.22,44 Additionally, the infection could also be established by contamination of wounds or skin abrasions with fluids from marine fauna and infected ornamental fish.45 In this work, the joint application of bacteriological and molecular techniques allowed to establish the aetiology of the infection. Among the molecular targets used, the hsp65 gene is present in all mycobacteria and other related microorganisms such as Nocardia sp and actinomycetes, and it encodes a family of antigenic 65 KDa heat shock proteins that are expressed in stress situations for the microorganism. Although the gene is present in all mycobacterial species, gene sequences amplified as fragments of 439 and 294 bp by PCR are polymorphic enough to accumulate species-specific variations in their sequence, which have been exploited to reveal the genetic polymorphism. Due the polymorphism of the 439 bp region of the hsp65 gene has been the most used, it has accumulated the sequence most used worldwide for the NTM identification. Despite the bacteriological tests performed on the purulent secretion from patient´s lesions support the result of M. marinum as responsible for the infection, molecular analyses performed on the 439 bp fragment amplified from hsp65 gene showed some ambiguities on the identification. In this regard, while the restriction analysis of the 439 bp amplicon was capable to rule out M. kansasii type 1, the polymorphism accumulated in this sequence was incapable to reveal differences among restriction patterns of clinical samples, M. marinum type 1 and M. malmöense type 2, as it can be deduced by the values of Euclidean distances calculated. A feasible explanation to this result could be attributable to the error of technique. In this regard, the restriction patterns of mycobacteria present in samples from patient differ from those of the reference strains by an average of 5 bp, and the estimation of the molecular size of the fragments obtained by the technique has an error of ±5 bp.17
As a consequence of this, the confirmation of the bacteriological tests required the analyses of samples using other molecular targets such as the sequence of 294 bp from hsp65 gene and the 16S-23S non-transcribed intergenic spacer gene of rRNA, which determined restriction patterns totally compatible with M. marinum and incompatible with M. malmöense. Similar results and difficulties for the differentiation of M. marinum and M. malmöense using the hsp65 gene as molecular target was described by Nolte et al.47 in a case of cervical lymphadenitis of mycobacterial aetiology. According to their results, the restriction analysis of a fragment amplified by PCR from hsp65 gen pointed to M. malmöense as the aetiological agent, however sequence analysis of the hsp65 fragment and the complete sequence of the gene coding for the 16S subunit of the amplified rRNA allowed them to identify unambiguously the mycobacterium isolated as M. marinum, suggesting that previous results obtained with the hsp65 gene were attributable to a point mutation in the sequence of this gene. Normally the hsp65 gene of M. marinum lacks of a restriction site for the BstEII enzyme, but a point mutation on the 439 bp fragment of this gene can determine the appearance of a restriction site for this enzyme compatible with the polymorphic pattern of M. malmöense. Based on these findings, the authors suggest the need to include more than one method in the molecular identification of these species, and that the identification of these species of mycobacteria that only differ slightly in their genetic polymorphisms and patterns of susceptibility to drugs such as ethambutol and rifampicin is important. The exploration of different molecular targets for specific mycobacterial identification will become more frequent every day due to the increasingly well-documented role of NTM described as opportunistic pathogens. To this is added the increasing trend in the number of communications on the role of new species of mycobacteria in human and animal health.2,46 Additionally, the characteristics of the case lead us to suggest, as it has been indicated by O'Connor et al.7, that is very important to keep and continuing to use the bacteriological techniques as the first line of identification and to apply and include more specific and sensitive molecular tests that exploit different mycobacterial targets with the aim to broaden the specificity of the mycobacterial identification.
The appropriate confirmation of the mycobacterium species responsible for the clinical features of the case herein presented, required the use of more than one molecular target, specifically two from hsp65 gene and one sequence from the non-transcribed intergenic spacer of rRNA. In this regard, the analyses of the restriction patterns obtained with the 294 bp fragment from hsp65 gene and the non-transcribed intergenic spacer of rRNA, allowed the adequate and unambiguous molecular confirmation of mycobacteria such as M. marinum in concordance with the results of biochemical and physiological tests carried out independently, that resulted in the presence of M. marinum in LJ cultures.
The authors would like to thank the technical staff of the Bacteriology Section from the Institute of Tropical Medicine (UCV) for their invaluable work. This work was partly financed with resources from the National Science and Technology Fund of Venezuela (FONACIT) project N°980036542, and Misión Ciencia project G-2007001442.
The authors (Correa de Adjounian María Fernanda; Rossi S Marcello Salvatore; Chalbaud Adriana; María Gabriela Liscano; Peralta Andreina; González, Deyanira; Adjounian María Fernanda; Pérez-León Carlos.; González Francisco; Hernández Cohinta; González-Rico Susana; Gómez María) declare that there are no competing interests regarding the publication of this paper.

©2018 Correa, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.