eISSN: 2373-6372


Research Article Volume 13 Issue 3
1Khwarizmi Institute of science and technology, Qeshm island, Hormozgan, Iran
2Congress 60 Non-governmental organization, Iran
Correspondence: Arvin Haghighatfard, Khwarizmi Institute of science and technology, Qeshm island, Hormozgan, Iran
Received: May 09, 2022 | Published: May 26, 2022
Citation: Dezhakam H, Dezhakam A, Haghighatfard A. Taper up-off treatment of opium may reverse some dysregulations of gene expression patterns caused by four type of leukemia, AML, ALL, CML and CLL; a pilot animal model study. Gastroenterol Hepatol Open Access. 2022;13(3):88-92. DOI: 10.15406/ghoa.2022.13.00502
Leukemia is a group of lethal diseases characterized by hematological neoplasm with complex and abnormalities in proliferation of bone marrow stem cells, that cause sever loss of functional capacity of hematopoietic tissue. Four most common types of leukemia are included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL) and chronic myeloid leukemia (CML). It has been reported that opium besides of pain killer effects, may have positive effects on cardiovascular disease treatments. Opium may consider as a potential treatment method in cancer. The present study aimed to assess the gene expression pattern alterations in the rat models of four types of Leukemia (ALL, AML, CLL, and CML) under taper up-off treatment method with of opium.
Present study was included twelve group, referred to four type of Leukemia (ALL, AML, CLL, and CML) that each type studied in three group. Group one contented normal rats, group two rat model of each type of leukemia with no treatment and group three the rat model of that type of leukemia with 26 days period of opium taper up-off treatment. After the scarification of rats, the bone marrow tissue were extracted. Then, RNA extraction and cDNA synthesis had been conducted on bone marrow tissues. Whole genome expression profiling was conducted by using the Affymetrix GeneChip Array Platform. Microarray results were confirmed by Real time PCR. Microarray analysis detected that hundreds of genes which were involved in Ontogenesis and apoptosis, humoral immune system, tool like receptors, natural killer cells regulators and metabolic pathways and were deregulated in leukemic models, were partially reregulated in treatment groups. Results showed that opium may have positive effects on reregulation of affected gene expression patterns in all types of leukemia. It may suggest that opium taper up-off treatment could be consider as re-regulator of immune system and a novel potential treatment for leukemia with low side effects and high viability.
Keywords: leukemia, opium, taper up-off, microarray
Leukaemia or leukemia is a group of blood cancers which mostly begin from the bone marrow and lead to production of several abnormal blood cells called leukemia cells.1 Bleeding, bone pain, fatigue, fever, vulnerability to infections are main symptoms of leukemia. Besides of rare types of leukemia, four most common and most lethal types of leukemia are included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL) and chronic myeloid leukemia (CML).2 These four major types account for 85% of all leukemia. CLL is the most common, but AML accounts for ~42% of all leukemia deaths. Most common type of cancer in children, is acute lymphoblastic type, on the other hand more than 90% of all adult subjects of leukemia, are diagnosed for CLL and AML.3 Worldwide prevalence of leukemia is approximately 2.43 million (95% UI 2.19 million to 2.59 million) with an age-standardized prevalence rate (ASPR) of 32.26 (95% UI 29.02 to 34.61) per 100,000 population with about the 400,000 death per year all around the world. Diagnosis of leukemia is typically is based on clinical symptoms, blood tests bone marrow biopsy and cytogenetic examinations.4
While the etiology of leukemia is not completely clarified, a combination of heritable genetic factors including mutations and chromosomal abnormalities like Down syndrome and epigenetic changer environmental factors such as smoking, radiations, several chemical compounds, prior chemotherapy, etc. are reported as risk factors of leukemia.5 Different types of chemotherapeutic drugs, radiation therapy, targeted therapy, and bone marrow transplant, alone or in combination with each other are currently using for treatment of leukemia.6 In addition, the Immunotherapy which prompts the immune system to identify and destroy cancerous cells is most novel treatment for leukemia. Despite of improvement in treatment success in the developed world, Five-year survival rate is lower than 65% in the United States and about 30–40% worldwide.
This disappointing survival rate of leukemia As well as considerable rate of resistance to chemotherapy, side effects of radiation and specificity of immunotherapy that made it unavailable for many cases based on their molecular testes results, has inspired much work on the development of new treatment methods. During last decades, several studies have revealed novel insights to treatment of leukemia. These treatments should be tested in animal models, and that’s' why the impact of animal models of cancer on leukemia research is very important.7
Opium is the dried latex obtained from the seed capsules of the "Papaver Somniferum" or opium poppy which has been a source of Morphinan-based painkillers with risks of addiction.8 Opium is effective in the treatment of different kind of acute and chronic pain that traditionally was used worldwide.9 On the other hand opium abuse and opium dependence is a major social and health problem all around the world.10 It has been reported that opium affects several mechanisms in body including the central nervous system (CNS), immune system, kidney functions, respiratory and cardiovascular systems.11,12
Opium effects on mRNA level of mammalian genes is mostly unclear. Opioid agents have major role in reduction of pain in cancer patients while the role of opiate use on epigenetic, immunological and hematological parameters as well as various metabolic and biological processes, in cancer is not clarified. Previous studies determined that the potential tumor-promoting, proliferation and migration effects of opiates are contradictory, and both growth-promoting and anti-tumor effects have been observed.13 That's why that shedding light on complexity of opium role in cancer treatment processes is tremendously important.
In Present study effect of opium taper up-off treatment in a laboratory designed method to expression profiling of rat models of four most lethal types of leukemia, ALL, AML, CLL, and CML were assessed. We aimed to understand that how the orally consumption of opium in a bell shaped dose curve could altered the several gene expression patterns which were previously affected by leukemia and is this alteration may reduce the severity of the disease of not.
Animal modeling of leukemia
To achieve the rat models of leukemia, hematopoietic stem cells (HSC) purified from patients' driven bone marrow human cell lines for each leukemia types (AML,ALL,CML,CLL). Human leukemic cells had been directly injected into immunocompromised and lethally irradiated rats. To increase the survival rate of host rats beyond this time point, lethally irradiated recipient rats were transplanted with mixtures of MHC-I matched rescue BM plus MHC-I-different BCR-ABL1 transduced BM. Successfulness Xenograft models assessed by pulmonary hemorrhage, splenomegaly and increased numbers of mature granulocytes in peripheral blood. Upon disease onset in rats the treatment was began. Animal modeling were performed based on previous studies.14 The study was included 13 groups of rats. One group was consisted of normal rats with no cure, act as control group. In rest of 12 groups, each three groups were used to study of one type of leukemia including rat model of that type of leukemia without treatment and two group of rat model of that type of leukemia with 26 days period of opium taper up-off treatment with two different starting dosage of opium. Each group was including six male rat (aged 35 days and weighing 200-250 grams). List of groups and descriptions of each group were presented in Tables 1-3.
Number |
Group name |
Description of group |
1 |
A |
rat model of ALL with no treatment |
2 |
B |
rat model of ALL with DST1 treatment |
3 |
C |
rat model of ALL with DST2 treatment |
4 |
D |
rat model of AML with no treatment |
5 |
E |
rat model of AML with DST1 treatment |
6 |
F |
rat model of AML with DST2 treatment |
7 |
G |
rat model of CLL with no treatment |
8 |
H |
rat model of CLL with DST1 treatment |
9 |
I |
rat model of CLL with DST2 treatment |
10 |
J |
rat model of CML with no treatment |
11 |
K |
rat model of CML with DST1 treatment |
12 |
L |
rat model of CML with DST2 treatment |
13 |
M |
Control group including normal rats with no cure |
Table 1 Name of all thirteen groups with Description of each group listed in this table. Each group was including six male rat (aged 35 days and weighing 200-250 grams)
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic myeloid leukemia; CML, chronic lymphocytic leukemia; DST1, Dezhakam-step-time treatment method with 1 mg/mL as first day dosage; DST2, Dezhakam-step-time treatment method with 2 mg/mL as first day dosage
Taper up-off treatment with opium
Opium tincture used to force-feeding to rats. In this laboratory designed method of taper up-off treatment called Dezhakam-step-time (DST) method, Opium tincture dosage start from the lowest dose and the dose will increase with 20 % rate for each day, until 13 days. Then the dosage of feeding reduce by the same 20% rate for next 13 days, and the first day of these 26 days and the final day will take the same dosage. Two starting dosage were used in two group of treatment for each type of leukemia. The first starting dosage in one treatment groups was 1 mg/mL (called "DST1") and first starting dosage in the other treatment group was 2 mg/ml (called "DST2").
Bone marrow sampling and RNA extraction
Bone marrow tissues of sacrificed rats were collected. Samples were put on process of RNA extraction. RNA was extracted from samples immediately after samplings according to standard protocols using by RNA Purification kit (GeneJET™ RNA Purification Kit#K0732, Thermo scientific - Fermentas, Latvia). Genomic DNA contamination removed from extracted RNA using DNase Treatment & Removal Reagents (DNase I, RNase-free (#EN0521) Fermentas, Latvia), according to the manufacture protocol. The quality and quantity of RNA were evaluated by Agarose gel electrophoresis and Nanodrop-1000 equipment respectively.
Synthesis of cDNA
Copied DNA synthesis was conducted by Transcription First Strand cDNA Synthesis Kit (RevertAid Premium First Strand cDNA Synthesis Kit #K1652, Thermo scientific -Fermentas, Latvia) according to manufacturer’s protocol.
Microarray assessments
Quality and integrity of extracted RNAs examined by Agilent 2100 Bioanalyzer (Agilent Technologies) before the beginning of the microarray process. Gene expression profiling analyses were examined using the Affymetrix GeneChip™ Rat Genome 230 2.0 Array that is the first whole genome tool, suitable for Oncogenes, toxicology, neurobiology, and other researches using rat models. Labeling and fragmentation of aRNA targets, followed by hybridization, and scanning based on manufacturer's protocol (Affymetrix Santa Clara, CA). Total RNA (100 Nano gram for each sample) was processed by the GeneChip 3′ IVT Express Kit. RNAs were reverse transcribed and converted to double-stranded cDNA prior to biotin labeling during in vitro transcription. Then fifty micrograms of labeled aRNA were fragmented, and quality control (QC) was evaluated using the Agilent Bioanalyzer. Fragmented aRNA was hybridized on Affymetrix GeneChip™ Rat Genome 230 2.0 Array for sixteen hours at 45°C. Then, arrays were washed and stained using the GeneChip Hybridization, Wash, and Stain Kit on the GeneChip Fluidics Station 450. Finally, chips were scanned by the Affymetrix GeneChip Scanner 3000, and all arrays passed the QC criteria examination.
The GeneChip analysis was performed in Genesis 2.0 (Gene Logic Inc.) and with Microarray Analysis Suite (MAS) 5.0, Data Mining Tool 2.0, and Microarray Database software (available at http://www.affymetrix.com). All represented genes in the GeneChip, had been normalized and scaled to hundred signal intensity. Filtering conducted for false-positive results reduction with MAS 5.0. Then, passed genes were analyzed by Genesis 2.0 (GeneLogic Inc., Gaithersburg, MD, USA) and DAVID software (Strand Genomics, Redwood City, CA, USA). Analysis of variance test (ANOVA) was used for all probe sets to determine significantly changed expressed genes. ANOVA testing for differentially expressed genes were followed by post hoc t-test for evaluation of contrasts groups one by one for identification of differences between groups. Ratio greater than two fold and the significant corrected P values after Bnferroni multiple testing correction were criteria to consider the gene as differentially expressed gene.
Confirmation with Real-time PCR
Top ten most differentially expressed genes in each microarray analysis had been confirmed with Real time PCR. Specific primers and probes were designed using "oligo7" software and were blasted on the NCBI website. Serial dilutions (1: 4) of pooled cDNA from total extracted RNA of randomly chosen normal rat samples used to draw the standard curves. CFX96 Touch Real-Time PCR Detection System (BIO-RAD, California, United States) used for triplicate method Quantitative Real Time-PCR. The R2 value more than 0.99 in the standard curve and no signal for no-template control samples were two quality checking criteria of qPCR assessments. Efficacy of PCR reaction calculates using online software of Lin-Reg PCR (Amsterdam, Netherland). Real time PCR was performed by TaqMan® PCR Starter Kit, Thermo scientific - Fermentas, Latvia). Livak formula was used to ratio calculation.
Enrichment pathways and gene ontology assessments
Comprehensive enriched functional examinations was performed on differentially expressed genes list for each study group. We used enrichment algorithms integrated into the online Database for Annotation, Visualization, and Integrated Discovery (DAVID 6.8 version) to functional annotation and gene ontology analysis. On the other hand an online data base, Kyoto encyclopedia of genes and genomic (KEGG) pathway enrichment tool used for molecular pathway mapping and gene ontology analysis on differentially expressed genes (DEGs). Gene ontology terms and KEGG pathways with corrected P values were reported and listed to use for visualization as Venn diagram with online software of Van de Peer Lab Bioinformatics and Evolutionary Genomics.
Statistical analysis
Statistical analysis analyzed by SPSS, version 25. Kolmogorov-Smirnov test used to normal distribution evaluation of variables. One way ANOVA analysis was used for statistical differences in multiple group comparisons. RNA integrity number, cDNA synthesis quality, plates/runs of qPCR, and primer efficiency were added as covariates, and persistence of the significant difference between groups was examined by ANCOVA to control any potential confusion. Bonferroni correction was used for multiple comparisons corrections. Descriptive data are expressed as mean ± SD.
Results of gene expressions of microarray assessments and differentially expressed genes in each group were presented table 2. In no cure groups, mostly affected gene expressions detected in CLL and ALL, CML and AML were in the next ranks. Significant association between in groups under DST1 (p=0.0001) and DST2 (p=0.0001) treatments increase of opium dosage and reduction of total DEGs especially up-regulated genes were detected.
Number |
Group name |
Total DEGs |
Up-regulated genes |
Down-regulated genes |
1 |
A |
2883 |
2097 |
786 |
2 |
B |
1622 |
1180 |
442 |
3 |
C |
820 |
510 |
310 |
4 |
D |
1820 |
1332 |
488 |
5 |
E |
1062 |
743 |
319 |
6 |
F |
540 |
320 |
220 |
7 |
G |
3257 |
2363 |
894 |
8 |
H |
1559 |
1033 |
526 |
9 |
I |
809 |
607 |
202 |
10 |
J |
2106 |
1573 |
533 |
11 |
K |
1277 |
941 |
336 |
12 |
L |
516 |
299 |
217 |
Table 2 Differentially expressed genes results of microarray assessments in comparison of each group with group M (control group)
A, rat model of ALL with no treatment; B, rat model of ALL with DST1 treatment; C, rat model of ALL with DST2 treatment; D, rat model of AML with no treatment; E, rat model of AML with DST1 treatment; F, rat model of AML with DST2 treatment; G, rat model of CLL with no treatment; H, rat model of CLL with DST1 treatment; I, rat model of CLL with DST2 treatment; J, rat model of CML with no treatment; K, rat model of CML with DST1 treatment; L, rat model of CML with DST2 treatment; M, Control group including normal rats with no cure
Top 10 genes with most differentially expression in microarray examinations were selected for Real time PCR confirmation. Findings were indicated that Real time PCR results confirmed the results of microarray analysis in all comparisons between groups due to direction of expression. Also no significant difference was found between expression alteration level calculated by ratio analysis between microarray results and Real time PCR results. Enrichment analysis showed numerous of molecular pathways were altered in all types of leukemia rat models. In groups under DST treatments, the affected and dysregulated pathways were significantly reduced in DST1 treatments in all types of leukemia. In addition, in ALL, AML and CLL groups, significant reduction of affected pathways determined in DST2 treated groups compared with DST1 treated groups. Besides of tumor suppressor genes and proto-oncogenes, most of the affected pathways in CML were involved in T helpers, MAPK signaling, transcription factors related to interleukin genes and metabolic pathways; in CLL were involved in cell adhesion, cell signaling and transcription factors ; in AML were involved in immune system, T helpers, MAPK signaling and metabolic pathways; and in ALL were involved in humoral immune system, tool like receptors, natural killer cells regulators and metabolic pathways. Statistical data of pathway analysis between groups in each leukemia type were presented in table 3. Comparisons of dysregulated molecular pathways number between each groups in each type of leukemia were presented as Venn plots in Figure 1 to 4.
Number |
comparisons |
p-value |
1 |
A vs. B |
0.0003 |
2 |
A vs. C |
0.0002 |
3 |
B vs. C |
0.008 |
4 |
D vs. E |
0.0004 |
5 |
D vs. F |
0.0002 |
6 |
E vs. F |
0.006 |
7 |
G vs. H |
0.0003 |
8 |
G vs. I |
0.0001 |
9 |
I vs. H |
0.007 |
10 |
J vs. K |
0.0003 |
11 |
J vs. L |
0.0002 |
12 |
K vs. L |
0.04 |
Table 3 Comparisons of pathway analysis between groups in each three groups of each leukemia type
A, rat model of ALL with no treatment; B, rat model of ALL with DST1 treatment; C, rat model of ALL with DST2 treatment; D, rat model of AML with no treatment; E, rat model of AML with DST1 treatment; F, rat model of AML with DST2 treatment; G, rat model of CLL with no treatment; H, rat model of CLL with DST1 treatment; I, rat model of CLL with DST2 treatment; J, rat model of CML with no treatment; K, rat model of CML with DST1 treatment; L, rat model of CML with DST2 treatment; M, Control group including normal rats with no cure
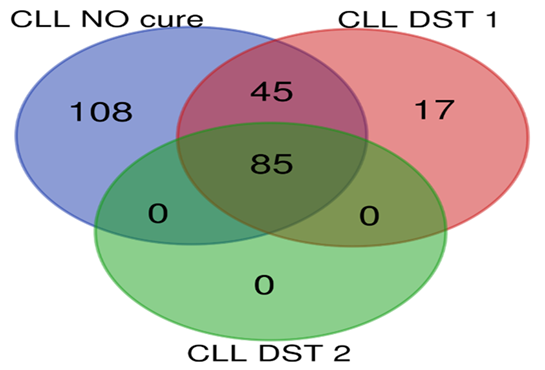
Figure 3 Pathway analysis for CLL groups; group G: CLL NO cure (blue circle), group H: CLL DST1 (red circle), group I: CLL DST2 (green circle).
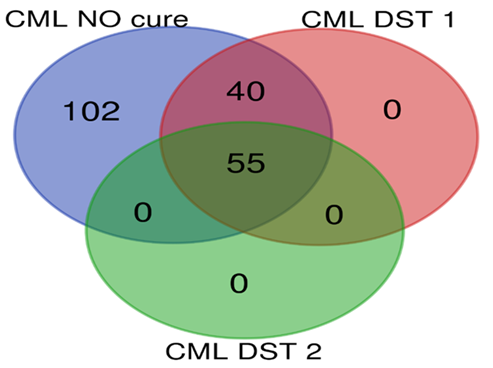
Figure 4 Pathway analysis for CML groups; group J: CML NO cure (blue circle), group K: CML DST1 (red circle), group L: CML DST2 (green circle) .
Leukemia is a group of lethal cancers that affect the blood, bone marrow, and lymphoid system, known as tumors of the hematopoietic and lymphoid tissues.15 Results of our study showed that DST method for increasing and decreasing the dose of opium can help to regulate the pattern of altered genes expressed in different types of leukemia. The mechanism and mode of action of the DST method at the molecular level is not yet known, but it seems that opium, with several alkaloids, can help to regulate upstream mechanisms of gene expression and epigenetics, especially transcription factors.
Opium has been prescribed for patients as painkiller due to its analgesic, hypnotic, antitussive, and antidiarrheal effects for many years. In the other hand, opium as an addictive and psychoactive drugs is the cause of addiction of about 16.5 million individuals' worldwide.16 Beneficial or impartial effects of opium in the cardiovascular system were reported in previous studies and consider opium as an immunosuppressive factor and reveal an increase in inflammatory mediators like C-reactive protein (CRP), interleukin-17, and interleukin-1 associated with chronic opium.17
The effects of opium on immune system function are not clarified. Previous studies reported inhibitory effects on immunosecretion and signaling pathways involved in maturation and function of immune cells.18 It seems that to interpretation of opium effects on immune system, the dosage and duration of exposure is quite critical. While acute and high dosage of opioids may affect the innate and acquired immune systems and inhibit the immune cells differentiation, chronic morphine exposure is associated with opioid receptor down-expression in neural cells, and overexpression in immune cells. Up-regulation of opioid receptors are clearly associated with increased Th2 T-helper cell differentiation.19 Some reports suggested that long-term opioid use may increases hypothalamus-pituitary-adrenal axis activity that leads to increased production of glucocorticoids, increasing the release of neuropeptide Y and down regulation of natural killer (NK) cells cytotoxicity.20
It seems that inflammation, regarding of its major role in development and progression of different types of cancer, especially leukemia has been forgotten in many researches about opioid effects on cancer and immune system as well. For example development of specific inhibitors of PI3K as an inflammatory associated agent is considered as a new treatment method for hematopoietic malignancies as well as for inflammatory and autoimmune diseases.21 It is well known that primary response to inflammation is a dramatically increase in survival of the target cell and promotion of maintenance of the cell. Mechanisms involving Bcl-2 family proteins and Pim family proteins which have been discovered in several hematological malignancies especially leukemia. Multiple mechanisms related to inflammation exists which result in increase in transcript and protein level of anti-apoptotic gene families, as well as induction of post-translational modifications and phosphorylation that lead to stability of pro-survival proteins and inhibits pro-apoptotic protein activity. Several small molecules pharmacological inhibitors are in early clinical trials for patients with hematological malignancies.22
Interestingly mesenchymal Inflammation cause genotoxic stress in hematopoietic stem cells that cause development of several gene expression alteration in human Pre-leukemic cells.23 inflammatory signaling induce genotoxic stress in heterotypic stem and progenitor cells that means the reregulation of inflammatory pathways could be consider as a new way to treatment of leukemia or strength the immune system to fight against the cancer cells. The significant reduction of cytokines and interleukins were detected in all four type of leukemia in both DST1 and DST2 group compared to leukemic models with no treatment groups. Human studies about the effects of air pollutions such as PM2.5 exposure, confirmed that enhanced expressions of inflammatory cytokines could act as major cause of promotion and progression of leukemia.24
Results of present study may raise this hope that if we can rich the golden dosage and accurate treatment protocol, we can use the anti-inflammatory effect of opium to inhibit the cell survival and anti-apoptotic activities in cancer cells especially in hematological malignancies. It seems that these anti-inflammatory effects in turn may lead to re-regulation of hundreds of molecular pathways that affected by leukemia and it may reduce the severity of disease as well as help to immune system to kill the cancer cells.
None.
The authors declare no conflicts of interest.

©2022 Dezhakam, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.