eISSN: 2373-6372


Research Article Volume 14 Issue 4
1Department of Dermatology, Case Western Reserve University, USA
2Department of Dermatology, The Center for Applied Health Sciences, USA
Correspondence: Mahmoud Ghannoum, Integrated Microbiome, A Cancer Resource Core, Case Western Reserve University and University Hospitals Cleveland Medical Center 11100 Euclid Avenue, Wearn 311, Cleveland, Ohio 44106-5028, Tel 216-844-8580
Received: June 26, 2023 | Published: July 14, 2023
Citation: Retuerto M, La-Monica MB, Ziegenfuss TN, et al. Probiotic supplementation of pea-derived protein alters the gut microbiome balance in favor of increased protein degradation, reflected in increased levels of essential amino acid in human plasma. Gastroenterol Hepatol Open Access. 2023;14(4):93-103. DOI: 10.15406/ghoa.2023.14.00553
Aim: The primary aim of this clinical study was to determine if dietary supplementation with the probiotic, BIOHM FX (BFX), altered the gut microbiome balance following ingestion of 15g pea protein (PP) and enhanced the absorption of non-animal proteins determined via quantification of essential amino acids (EAAs). Thus, we compared the effects of pea protein alone vs. pea protein + BFX on microbiome changes and plasma levels of EAAs.
Methods: A placebo-controlled crossover clinical study in active men (n=40) was performed during which quantification of abundance levels of gut bacterial and fungal (bacteriome and mycobiome) organisms were assessed. In addition, plasma EAAs were measured pre- and post- ingestion of the pea protein +/-BFX for 180 min. Stool samples were analyzed for changes in microbiome composition from baseline and compared for PP versus PP+BFX. Self-reported changes in gastrointestinal (e.g., bloating, flatulence) and quality of life (e.g., fatigue, mood, and energy) indices were also measured.
Results: Participants ingesting PP + BFX exhibited a distinct microbiome profile compared to baseline and ingestion of PP. Differences in plasma EAAs showed a trend for an interaction (P=0.097) and post hoc testing at 120 min showed a significant difference (P=0.047) between PP and PP+BFX. Microbiome analysis of stool samples showed that the pathogens Escherichia coli, Prevotella copri, Shigella flexneri, and Brevundimonas diminuta were lower in PP+BFX compared to PP. The abundance of Candida albicans was lower and the level of Saccharomyces cerevisiae was higher in PP+BFX compared to PP. Interestingly, the abundance of Pseudomonas species, cyanobacteria phyla and the fungal species Galactomyces geotrichum were elevated when the combination of PP+BFX were consumed by study subjects (P<0.05). Other than main effects of time there were no significant differences between treatments in self-reported gastrointestinal (GI) and well-being markers.
Conclusion: Our results indicate that the addition of BFX to PP alters the gut microbiome composition, aiding in the absorption of dietary non-animal proteins and increasing essential amino acid appearance in plasma.
Keywords: probiotic, gut health, amino acid absorption, microbiome
Increased interest in potential sources of non-animal proteins, whether due to environmental concerns, worldwide overpopulation, or increased awareness of food allergens, has resulted in greater attention being given to a variety of plant proteins. Plant proteins offer a sustainable, lower-cost alternative to animal proteins. Although soybeans are a major food source produced in North America, field peas (Pisum sativum) account for nearly 26% of the worldwide production of edible seeds.1 Two commercial protein supplements (whey and pea protein powders) are popular with athletes, vegetarians, and vegans. Both whey and pea protein powders are considered complete proteins, as they contain all nine EAAs, though pea protein has very small amounts of methionine.2 Importantly, while many types of whey products contain lactose and/or gluten allergens, pea proteins are free from the most common food allergens. Thus, these supplements could serve as an invaluable dietary source of easily renewable protein. Combining these protein supplements with beneficial probiotics is an upcoming area of interest.3
The role of the gut microbiome regarding mechanism(s) of nutrient absorption has been extensively studied in recent years. Several studies have shown that various probiotics are capable of increasing essential amino acid absorption. 4,5,6 Production of enzymes by beneficial gut microbes have been postulated to facilitate absorption of micronutrients such as vitamins, minerals, and amino acids through the mucosal layer of the small intestine, made up of enterocytes and mucin secreted by goblet cells.7–9 However, disruption of this mucin layer, as occurs during dysbiosis of the gut microbiome, may enable invasion by pathogenic organisms, resulting in formation of thick biofilms which prevent nutrient absorption.10
Previously, our analysis of the gut microbiome of patients with Crohn’s disease (CD) showed elevated levels of the pathogens Candida tropicalis, Escherichia coli, and Serratia marcescens compared to their healthy family members.11 We subsequently demonstrated that the combination of these three organisms resulted in the production of robust biofilms in vitro and in a murine in vivo model.10 Further studies using an in vitro biofilm model showed that the novel probiotic formulation BIOHM FX (BFX) (BIOHM Health, LLC, Cleveland, OH), consisting of Saccharomyces boulardii 16mxg, Lactobacillus acidophilus 16axg, L. rhamnosus 18fx, and Bifidobacterium breve 19bx strains in combination with the enzyme amylase, significantly reduced the thickness of polymicrobial biofilms.12
Considering the in vitro activity of BFX on biofilm inhibition and thickness reduction, we hypothesized that BFX should enhance the absorption of micronutrients in the small intestine. Thus, the passage of vitamin C and casein, as representative of vitamins and proteins, respectively, through an epithelial monolayer was tested with and without the addition of BFX supernatant. Results showed that BFX significantly increased the permeability of both vitamin C and casein (P values <0.05 and 0.0001, respectively) through the Caco-2 cell monolayer overlaid with polymicrobial bacterial-fungal biofilms, elicited by C. tropicalis, E. coli, and S. marcescens exposure.13
Given the results demonstrating that BFX could reduce biofilm formation, leading to increased nutrient absorption in vitro, we wondered whether or not BFX would also enhance pea protein absorption in subjects consuming pea-derived protein supplements. A human clinical study was designed to compare the effects of a pea protein absorption in the presence or absence of BFX. Although several commercial supplements are considered complete proteins (e.g., whey and pea powders) that contain all nine essential amino acids, pea protein was chosen as the supplement because of its non-allergenic properties, as opposed to whey powder, which may contain lactose and/or gluten allergens.2
The aims of this clinical study were to: a) compare the effects of pea protein alone vs. pea protein + BFX on the microbiome composition of participating subjects using next generation targeted sequencing of bacteriome and mycobiome microbiota, b) compare the effects of pea protein alone vs. pea protein + BFX on plasma essential amino acid (EAA) levels, c) compare the effects of pea protein alone vs. pea protein + BFX on self-reported visual analog scales (VAS) for gastrointestinal bloating, flatulence, fatigue, mood, and energy, and d) assess safety and tolerability as determined by vital signs and adverse events.
Our hypothesis was that PP+BFX would alter the gut microbiome profile differentially from consumption of PP, and that the addition of BFX would lead to increased appearance of plasma EAA while also improving gastrointestinal (GI) tolerance.
Clinical study design
The current study was registered on ClinicalTrials.gov (ID number NCT05657314). This double-blind, randomized, placebo-controlled, crossover clinical trial was designed to solely recruit recreationally active men (n=40). A consort diagram of the study design and enrollment features is shown in Figure 1. The subjects were recruited at a single investigational center in Ohio (The Center for Applied Health Sciences). Following informed consent, eligible subjects were randomly assigned in a 1:1 ratio to one of two study arms: 15g of Yantai Shuangta 85% pea protein with placebo (cellulose) dissolved in 12 fl. oz. of water (PP alone) or 15g of Yantai Shuangta 85% pea protein with 1 billion colony forming unit of BFX (PP+BFX) dissolved in 12 fl. oz. of water. Enrolled subjects ingested the assigned daily supplement for 4weeks, followed by a 1-week washout period (absent of any supplementation). Subjects were then given the alternate supplement for an additional 4 weeks. This crossover approach enabled each subject to serve as his own control, thus reducing inter-subject variability and enhancing statistical power.
Participants
40 healthy men completed all study visits, but 39 subjects were analyzed (See Table 1 for subject characteristics). All participants were in good health as determined by physical examination and medical history, recreationally active men (exercise ≥2-3d/wk for at least 1 year), between the ages of 18 and 55 years, and had a body mass index (BMI) of 18.5-29.9kg•m-2. Prior to participation, all participants indicated their willingness to comply with all aspects of the experimental and supplement protocol. Participants were excluded if they: (a) had a history of diabetes or pre-diabetes; (b) had a history of malignancy in the previous 5 years except for non-melanoma skin cancer (basal cell cancer or squamous cell cancer of the skin); (c) had prior gastrointestinal bypass surgery; (d) known gastrointestinal or metabolic diseases that might impact nutrient absorption or metabolism (e.g. short bowel syndrome, diarrheal illnesses, history of colon resection, gastro paresis, Inborn-Errors-of-Metabolism); (e) had any chronic inflammatory condition or disease; (f) had a known allergy to any of the ingredients in the supplement or the placebo; (g) had currently been participating in another research study with an investigational product or have been in another research study in the past 30 days; (h) had excessive caffeine intake (>600 mg) per day; (i) used corticosteroids or testosterone replacement therapy (ingestion, injection, or transdermal); (j) had ever been diagnosed with liver, renal, cardiovascular, or other metabolic disease; (k) consumed more than 2 standard alcoholic drinks per day (or more than 10 drinks per week) or had a history of drug abuse/dependence; (l) use of any prescription medications (particularly antibiotics and/or anti-inflammatories), or probiotics within the past 2 months; (m) current smokers; (n) had any other diseases or conditions that, in the opinion of the medical staff, could confound the primary endpoint or place the participant at increased risk of harm if they were to participate; or (o) did not demonstrate a verbal understanding of the informed consent document.
Participants were instructed to follow their normal dietary habits throughout their participation in the study. Participants were required to complete a 24-hour diet record prior to arriving at the laboratory for their first initial screening visit. Participants were given a copy of this dietary record and instructed to duplicate all food and fluid intake 24 hours prior to their subsequent laboratory visits. Prior to each subsequent visit participants were asked to verbally confirm their 24-hour prior diet adherence and ensure they had a normal night’s rest. In addition to replicating food and fluid intake for 24 hours prior, study participants were also asked to refrain from exercise for 72 hours prior, refrain from alcohol and caffeine 24 hours prior, and arrive 8 hours fasted to all testing sessions which were all verbally confirmed at the beginning of each study visit.
Clinical study visits
The study design and schedule of visits is shown in Figure 2. At Visit 1 (Screening), medical history and 24hr dietary recall were collected, along with routine safety blood work (CBC, CMP, and lipid panel). Following Visit 1, subjects underwent a baseline visit (Visit 2) consisting of body weight, blood pressure, heart rate, and a 3-day diet record. A baseline stool sample was provided at Visit 2 and collected at home prior to participant’s initial supplementation and sent for microbiome analysis which included bacterial (bacteriome) and fungal (mycobiome) abundance quantification. Participants were provided with their first assigned treatment on Visit 2 and instructed to consume the investigational product after their stool sample was collected. After 4 weeks of daily consumption, participants came in for post testing (Visit 3) where they ingested their final dose of the product in the presence of the research staff. Three days prior to Visit 3, participants were instructed to collect another stool sample and send it out for analysis. Visit 3 consisted of several procedures; 1) blood was drawn before and 30, 60, 90, 120, 180 min post-ingestion of the protein supplement (either PP alone or PP+BFX) for EAAs; 2) perceived changes in GI flatulence, GI bloating, level of fatigue, overall mood, and level of energy were assessed using a 100 mm anchored VAS (visual analog scale) before supplement ingestion and 60, 120, 180 min post-ingestion, 3) vitals were recorded before and 60, 120, 180 min post-supplement ingestion; 4) a 3-day diet record was performed prior to each visit; and 5) stool samples were collected at home and sent for microbiome analysis. At the end of Visit 3, participants were provided with their second assigned treatment and told to begin consumption of the investigational product after a one-week washout period where none of the investigational products were consumed. A sufficient washout period (one week) was then performed to allow complete digestion of the consumed assigned treatment prior to switching to the second phase of treatment.14 Again, 4 weeks later participants came in for post-testing (Visit 4) which included the same testing measures that were conducted at Visit 3. Participants were also instructed to collect another stool sample within three days prior to Visit 4 and send it out for analysis. The VAS provides a simple, reliable measure that is easy to follow and requires little time to complete. Participants can respond on continuous lines rather than Likert-type scales which allows them to rate their answer with little bias and any desirable amount of discrimination.15 The validity and reliability of VAS to assess fatigue and energy have been previously established.15
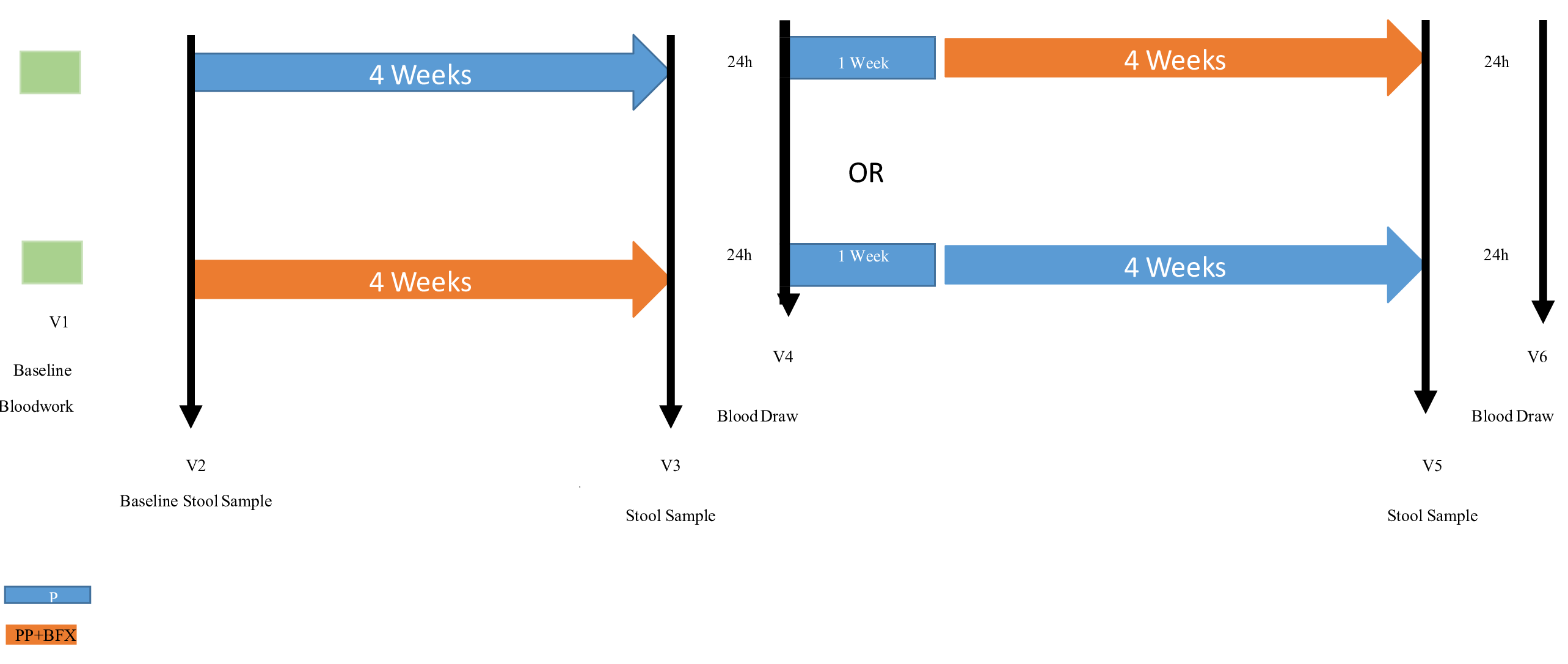
Figure 2 Shows the study design for this open label cross over study indicating the timing of the supplements, the samples obtained, the outcome measures, and the visit timing.
The study was conducted following ICH-GCP guidelines to ensure subject safety and scientific integrity of the data. Comprehensive side effect profile/adverse event monitoring took place throughout the study duration. There were no serious adverse events reported during the study. Mild adverse events were rare and included paresthesia and pre-syncope, associated with a muscle damage protocol (data not shown), rather than protein supplement ingestion. A table of adverse events is given in Supplemental Table 1.
|
|
Allocated Cross-Over (n=85) |
|
|
|
A (n=44) |
B (n=41) |
|
Severity |
|
|
|
Mild |
2 |
3 |
|
Moderate |
3 |
1 |
|
Severe |
|
|
|
Relationship to Study Procedures |
|
|
|
Unlikely |
|
1 |
|
Possible |
|
|
|
Probable |
5 |
1 |
|
Relationship to Test Article |
|
|
|
Unlikely |
4 |
1 |
|
Possible |
1 |
3 |
|
Probable |
|
|
|
Body System and AEs |
|
|
|
GastoIntestinal |
|
|
|
Bloating |
|
1 |
|
Flatulance |
|
1 |
|
Gastrointestinal Pain |
|
1 |
|
General Disorder |
|
|
|
Edema Face |
1 |
|
|
Nervous System |
|
|
|
Parathesia |
1 |
|
|
Pre-Syncope |
3 |
1 |
|
Total Number of Adverse Events Experienced During Study |
5 |
4 |
|
Total Number of Subjects Experiencing AEs: n (%) |
5/44 (11%) |
4/41 (10%) |
Supplemental Table 1 Summary of Adverse Events
Quantification of Essential Amino Acid (EAA) Concentration in plasma
Plasma amino acid concentrations were determined by liquid chromatography-mass spectrometry (LC-MS) using a QTrap 5500 Mass Spectrometer (AB Sciex, Foster City, CA, USA) and an internal standard method, as described previously. 16,17 The analytes were derivatized with 9-fluorenylmethoxycarbonyl chloride (Fmoc-Cl; 23186, St. Louis, MA, USA). Ions of mass to charge ratio of 340/144 for threonine, 338/116 for valine, 370/47 for methionine, 352/130 for isoleucine and leucine, 425/203 for tryptophan, 386/164 for phenylalanine, 598/154 for histidine, and 589/145 for lysine were monitored with selected ion monitoring on quadrupole one and three, respectively. Quantification of each peak was determined using MultiQuant software (version 2.1: AB Sciex).
Microbiome analysis of fecal samples
To determine the effect of supplementation with PP alone versus PP+ BFX on the microbiota profile, fecal samples were processed using our previously published methodology.18
DNA extraction
Fecal samples were analyzed for identification of their bacterial (bacteriome) and fungal (mycobiome) communities using Ion Torrent sequencing technology (Thermo Fisher Scientific, Waltham, MA, USA). Samples were transferred to tubes containing glass beads with the lysis solution included in the QiaAmpFast DNA Extraction Kit (QIAGEN, Germantown, MD, USA). Bacterial and fungal DNAs were isolated and purified following the manufacturer’s instructions with minor modifications. In this regard, we incorporated an additional bead-beating step (Sigma-Aldrich beads, diameter =500µm), with the MP FastPrep-24 speed setting of 6M/s and 2×40 s cycles. The quality and purity of the isolated genomic DNA were confirmed using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). DNA concentration was quantified using a Qubit 2.0 instrument applying the Qubit dsDNA HS Assay (Life Technologies, Carlsbad, CA USA) and adjusted to 100ng per sample. Extracted DNA samples were stored at −20º C until needed.
Bacterial 16S rRNA gene or pan fungal ITS amplicon library preparation.
For bacteria, the V3-V4 region of the 16S rRNA gene was amplified using 16S-515F: GTGCCAGCMGCCGCGGTAA and 16s-806R: GGACTACHVGGGTWTCTAAT primers, while the fungal ITS region was amplified using ITS1 (CTTGGTCATTTAGAGGAAGTAA) and ITS 2 (GCTGCGTTCTTCATCGATGC) primers. The reactions were carried out on a 100ng template DNA, in a 50µL (final volume) reaction mixture consisting of Q5 PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), for a final primer concentration of 400nM. Initial denaturation at 94º C for 3min was followed by 30 cycles of denaturation for 30 s each at 94º C, annealing at 57º C (16 s) or 59º C (ITS) for 30 s, and extension at 72º C for 10 s. Following the 30-cycle amplification, there was a final extension time of 15 s at 72º C. The size and quality of amplicons were screened on a 1.5% TAE agarose gels, separated using 100v, and electrophoresed for 45 min then stained with ethidium bromide. The PCR products were sheared for 20 min, using Ion Shear Plus Fragment Library Kit (Life Technologies, Carlsbad, CA, USA). The amplicon library was generated with sheared PCR products using Ion Plus Fragment Library Kit (<350 bp) according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). The library was barcoded with Ion Xpress™ Barcode Adapter and ligated with the A and P1 adaptors (Thermo Fisher Scientific, Waltham, MA, USA).
Next-generation sequencing, classification, and analysis
The adapted barcoded libraries were concentrated 4–6X in a speed-vac (Thermo Fisher Scientific, Waltham, MA, USA) and the concentrated pooled libraries were then quantified using a TaqMan Quantitation Kit (Thermo Fisher Scientific, Waltham, MA, USA). The libraries were adjusted to 100pM and attached to the surface of Ion Sphere particles (ISPs) using an Ion PGM Template OT2 400bp Hi-Q View Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions, via emulsion PCR.
The quality of ISP templates was checked using Ion Sphere™ Quality Control Kit (Part no. 4468656- Thermo Fisher Scientific, Waltham, MA, USA) with the Qubit 2.0 device. Sequencing of the pooled libraries was carried out on an Ion Torrent PGM System using the Ion Sequencing 400bp Hi-Q View Kit (Life Technologies, Carlsbad, CA, USA) for 150 cycles (600 flows) with a 318 v2 chip, following the manufacturer’s instructions. De-multiplexing and classification were performed using the Qiime Platform (ver. 1.8). The resulting sequence data were trimmed to remove adapters, barcodes, and primers during the de-multiplexing process. In addition, the sequence data were filtered for the removal of low-quality reads below the Q25 Phred score and de-noised to exclude sequences with a read length below 100bp.19 De novo OTU’s were clustered using the Uclust algorithm and defined by 97% sequence similarity.20 Classification at the species level was referenced using the Greengenes (v. 13.8) reference database 21 and taxa assigned using the nBlast method with a 90% confidence cut-off. 22 Abundance profiles for the microbiota were generated and imported into Partek Discover Suite v6.11 for principal components analysis (PCA). Diversity and correlation analyses and Kruskal– Wallis (non-parametric) analysis of variance were performed using abundance data and R statistical analysis software (CRAN, and Morgan) with packages (Psych and Vegan, Bioconductor). Diversity indices, including Shannon’s Diversity Index (SDI), Richness (N), and Pielou's evenness (PE), were calculated at all taxonomic levels.
Statistical analysis
Based on previous data, as well as similar studies in the literature, a sample size calculation was performed with an effect size of 0.25, significance level of 0.05, power of 80%, and a dropout rate of ~20%. With these parameters, a total sample size of 40 subjects was chosen for this study. The primary outcome variable was plasma EAA response, and the secondary variables were changes in gut microbiome composition and GI tolerance/GI health (flatulence, bloating, fatigue, mood, energy). Descriptive statistics (mean and SD) were used to quantify subjects’ physical characteristics. A table of demographics is shown for the study participants (Table 1). Two-way (group x time) linear mixed models were completed to assess group, time, and group x time interaction effects for EAAs and subjective rating for all VAS items. Paired samples t-tests or sidak post-hoc procedures were used to assess individual comparisons between time points and/or groups. SPSS and GraphPad Prism were used to perform these statistical analyses. For primary and secondary endpoints, post-hoc outcomes that indicated a significant difference (P value ≤ 0.05) or a trend (P value >0.51 to ≤ 0.1), Cohen’s D effect sizes were calculated to evaluate the magnitude of the observed effect between treatment groups. Change scores between baseline and 30, 60, 90, 120, 180min for EAA were also calculated. For microbiome data, statistical significance levels were calculated comparing the changes across groups by Welch’s t-test for a given genus, species, or phylum.
|
|
Total (N=39) |
|
Age (years) |
26.3 ± 7.9 |
|
Height (cm) |
179.7 ± 7.4 |
|
Weight (kg) |
83.7 ± 9.0 |
|
Body Mass Index (kg/m2) |
26.0 ± 3.0 |
|
Systolic Blood Pressure (mm Hg) |
122.3 ± 9.4 |
|
Diastolic Blood Pressure (mm Hg) |
74.5 ± 8.2 |
|
Resting Heart Rate (bpm) |
66.6 ± 9.8 |
Table 1 Demographics Summary
Quantification of Essential Amino Acid (EAA) concentration in plasma
All plasma amino acids displayed main effects of time (P≤0.001). A trend for a time by group interaction (P= 0.097) was noted for the plasma EAA temporal response. Post hoc testing indicated that PP+BFX had a significantly higher (P= 0.047; Cohen’s D =0.30) plasma EAA concentration (1,333.9 ± 271.2 µmol/L) at 120 min as compared to PP alone (1,277.3 ±2 58.3 µmol/L), a 4.4% increase. A trend for a time by group interaction (P = 0.066) was observed on the change from baseline for plasma EAA concentrations. Post-hoc testing indicated that PP+BFX had a trend (P= 0.081; Cohen’s D=0.36) for a relative increase in plasma EAA concentrations from baseline to 120 minutes post-ingestion as compared to PP alone (1,040 ±185.6 µmol/l vs. 996.7 ± 158.8 µmol/L, respectively) (Table 2). A trend for a time by group interaction (P=0.086) was observed on the change from baseline for plasma lysine concentrations. Post-hoc testing (data not shown) indicated that PP alone had a significantly greater (P= 0.029; Cohen’s D=0.36) increase in plasma lysine concentrations from baseline to 90 minutes post-ingestion as compared to PP+BFX (229.6 ± 171.4 µmol/l vs. 182.8 ± 188.5 µmol/L, respectively).
|
Variable |
Time* |
PP+ BFX |
PP alone |
|
EAA (µmol/L) |
0 mina |
1074 ± 164.2 |
1034.3 ± 180.2 |
|
30 minb |
1750.6 ± 450.6 |
1741.3 ± 432.4 |
|
|
60 minb |
1686.5 ± 465.2 |
1710.4 ± 442.6 |
|
|
90 minc |
1451.9 ± 331.6 |
1474.2 ± 320.0 |
|
|
120 mind,# |
1333.9 ± 271.2 |
1277.3 ± 258.3 |
|
|
180 mine |
1176.9 ± 257.2 |
1152.8 ± 217.5 |
Table 2 Plasma Essential Amino Acids
*Indicates a trend for an effect/interaction (P=≤0.10). # Indicates a trend between groups at that timepoint (p≤0.10). Times without the same letter are significantly different from one another (P=<0.05).
Subjective outcome variables and dietary records
There was a significant main effect of time (P≤0.001 – p=0.044) for flatulence, bloating, fatigue, mood, muscle tightness, and muscle soreness. There were no significant differences between groups or over time noted for energy (P>0.050). There were no differences over time or between groups in average calories, carbohydrates, fat, or protein (all P>0.05).
Microbiome analysis
Microbiome composition: stack plot
The supplementation of pea protein with BIOHM FX (PP +BFX) led to changes in the gut microbiota at the species composition level (Figure 3), particularly in bacterial and fungal species present in the gut at lower relative abundances, compared to both the baseline and PP alone. Similarly, subjects that received PP alone also exhibit patterns of microbiota change, largely in species of lower abundance, but to a lesser degree than PP + BFX (see Supplemental Figures 2A and 2B).
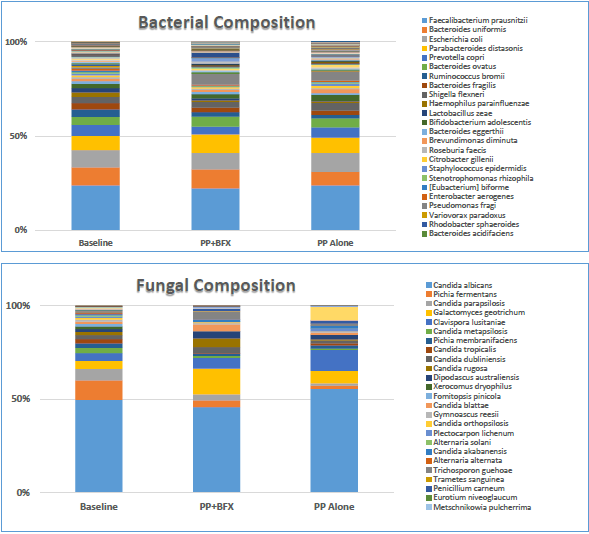
Figure 3 Microbial Composition: Stack Plot It shows the compositional differences in gut microbiota between treatment groups at the Species level. Relative Abundance of bacteria (16S-Bacteriome) and fungal (ITS-Mycobiome) composition is shown for the three treatment conditions (Baseline), Pea Protein + BFX (PP+BFX), and Pea Protein alone (PP alone). Microbiome Composition: Stack Plot of Bacteriome and Mycobiome.
Alpha diversity analysis
Among baseline and the two treatments analyzed (baseline, PP+BFX, and PP alone) a similar level of richness, SDI, and PE was observed at the phylum level (Table 3A&3B) in the bacteriome and mycobiome profile. The bacteriome changes in diversity were mostly in the PP alone cohort primarily observed as an increase in SDI. In contrast, PP alone contained the lowest richness. In regard to the diversity of the mycobiome, the PP+BFX treatment cohort exhibited a modest increase in species SDI with the greatest effect at genus level.
|
Bacteriome |
||||
|
Taxa |
Status |
Richness |
SDI |
Pilou's Eveness |
|
Phylum |
Baseline |
17 |
0.92 |
0.34 |
|
PP+BFX |
16 |
0.96 |
0.35 |
|
|
PP alone |
15 |
0.97 |
0.37 |
|
|
Genus |
Baseline |
170 |
2.32 |
0.46 |
|
PP+BFX |
171 |
2.33 |
0.46 |
|
|
PP alone |
160 |
2.39 |
0.48 |
|
|
Species |
Baseline |
92 |
1.97 |
0.45 |
|
PP+BFX |
94 |
2.10 |
0.48 |
|
|
PP alone |
89 |
2.14 |
0.49 |
|
Table 3A
|
Mycobiome |
||||
|
Taxa |
Status |
Richness |
SDI |
Pilou's Eveness |
|
Phylum |
Baseline |
2 |
0.13 |
0.22 |
|
PP+BFX |
2 |
0.18 |
0.27 |
|
|
PP alone |
2 |
0.11 |
0.19 |
|
|
Genus |
Baseline |
57 |
0.80 |
0.38 |
|
PP+BFX |
54 |
0.95 |
0.45 |
|
|
PP alone |
47 |
0.84 |
0.43 |
|
|
Species |
Baseline |
41 |
1.09 |
0.50 |
|
PP+BFX |
38 |
1.10 |
0.54 |
|
|
PP alone |
36 |
0.93 |
0.50 |
|
Table 3B
The microbiota biodiversity
The Venn diagram shown in Figure 4A illustrates the bacterial species level biodiversity richness data among the three treatment conditions (baseline, PP, PP+BFX), demonstrating that 302 species are shared across all comparator groups. An interrelationship between PP+BFX and PP shows that they share 9 common species. Interestingly, 4 unique species, Plantago atrata, Luteimonas mephitis, Pedobacter saltans, and Acidovorax caeni, are exclusive to PP+BFX. Comparing Baseline and PP+BFX, there are 5 shared species. The Baseline and PP alone groups share 4 species, with no unique species on either side, suggesting PP alone may be a subset of Baseline. Baseline contains 4 unique species, Prosthecobacter debontii, Weissella viridescens, Bdellovibrio bacteriovorus, and Clostridium difficile, not found in the other two groups, suggesting changes to the bacterial biodiversity by the supplementation of PP+BFX.
Figure 4B demonstrates the impact of the fungal species biodiversity among the three treatment conditions (baseline, PP, PP+BFX), with a total of 412 species shared across all groups. However, the biodiversity is primarily influenced by the PP+BFX cohort, which shares 43 and 42 species with the Baseline and PP cohorts respectively. Most striking is the presence of 71 unique fungal species within the PP+BFX treatment, significantly contributing to the overall biodiversity. Notably, 20% of these unique species are from the Penicillium genera, underlining a key influence of this genus within the unique fungal biodiversity profile of the PP+BFX treatment.

Figure 4 Venn diagram species level biodiversity richness for bacteriome (A) and mycobiome (B) among the three cohorts, it shows the overlap of bacteriome (A) and mycobiome (B) OTUs between.
Abundance microbiota analysis
A comparison of the microbiota composition of subjects at baseline versus PP+BFX shows that in PP+BFX treatment, Pseudomonas nitroreducens exhibited a significant increase in relative abundance, comparing baseline (0.0095%) with PP+BFX (0.0526%) (Table 4). This corresponds to a fold change of 5.5, (P = 0.02). Similarly, Pseudomonas stutzeri showed a significant (P = 0.037) 12-fold increase in relative abundance in PP+BFX (0.0704 OTUs) compared to baseline (0.0059). Another species belonging to the genera Pseudomonas, Pseudomonas umsongensis, increased in the PP+BFX cohort (P = 0.026), suggesting a substantial amplification of Pseudomonas following the intervention of PP+BFX.
|
Taxa |
|
Baseline RA (%) |
PP+BFXRA (%) |
Baseline Prevalence (%) |
PP+BFX Prevalence (%) |
PP+BFX: Baselin e: (FC) |
Elavated in |
Baseline: PP+BF X (p value) |
|
Bacterial Phyla |
Crenarchaeota |
0.0063 |
0.0139 |
71.1 |
73.7 |
2.2 |
PP+BFX |
0.048 |
|
Bacterial Genus |
Pseudomonas |
0.74 |
6.25 |
100 |
100 |
8.4 |
PP+BFX |
0.028 |
|
Alcanivorax |
0.0061 |
0.0018 |
55.3 |
31.6 |
-3.4 |
Baseline |
0.041 |
|
|
Brenneria |
0.0091 |
0.0010 |
31.6 |
15.8 |
-8.8 |
Baseline |
0.036 |
|
|
Dethiosulfatibacter |
0.00101 |
0.00018 |
21.1 |
7.9 |
-5.6 |
Baseline |
0.041 |
|
|
Bacterial Species |
Pseudomonas nitroreducens |
0.0095 |
0.0526 |
23.7 |
52.6 |
5.5 |
PP+BFX |
0.020 |
|
Pseudomonas stutzeri |
0.0059 |
0.0704 |
31.6 |
42.1 |
12.0 |
PP+BFX |
0.037 |
|
|
Acinetobacter rhizosphaerae |
0.0032 |
0.0128 |
15.8 |
28.9 |
4.0 |
PP+BFX |
0.046 |
|
|
Pseudomonas umsongensis |
0.0015 |
0.0068 |
13.2 |
26.3 |
4.5 |
PP+BFX |
0.026 |
|
|
Alcanivorax dieselolei |
0.0101 |
0.0035 |
47.4 |
21.1 |
-2.9 |
Baseline |
0.022 |
|
|
Tindallia Anoxynatronum |
0.00052 |
0.00002 |
21.1 |
2.6 |
-26.1 |
Baseline |
0.015 |
|
|
Fungal Genus |
Galactomyces |
3.1 |
10.1 |
68.4 |
78.9 |
3.3 |
PP+BFX |
0.048 |
|
Plectocarpon |
0.47 |
0.10 |
44.7 |
21.1 |
-4.8 |
Baseline |
0.037 |
|
|
Schwanniomyces |
0.0019 |
0.0003 |
21.1 |
10.5 |
-5.5 |
Baseline |
0.042 |
|
|
Fungal Species |
Galactomyces geotrichum |
4.1 |
12.0 |
68.4 |
78.9 |
2.9 |
PP+BFX |
0.046 |
|
Lasiodiplodia theobromae |
0.00008 |
0.00222 |
7.9 |
13.2 |
28.6 |
PP+BFX |
0.047 |
|
|
Plectocarpon lichenum |
0.70 |
0.15 |
44.7 |
21.1 |
-4.8 |
Baseline |
0.037 |
|
|
Stenocarpella maydis |
0.0002 |
0.0024 |
7.9 |
15.8 |
11.9 |
PP+BFX |
0.047 |
|
|
Umbilicaria lyngei |
0.0023 |
0.0002 |
15.8 |
5.3 |
-9.6 |
Baseline |
0.045 |
Table 4 Changes in Bacteriome and Mycobiome Composition Comparing Baseline to PP+BFX
In contrast, species such as Alcanivorax dieselolei and Tindallia Anoxynatronum show a decrease in relative abundance in the PP+BFX treatment group compared to the baseline. The reduction in these species may be related to either the pea protein or BFX exposure. However, because these species were not measured or found in PP alone, it cannot be distinguished whether it was the pea protein exposure or the BFX exposure that led to these changes. The presented data includes only those bacterial taxa that exhibited statistically significant changes as determined by a Welch's t-test (P < 0.05) and prevalence (>20%) for any single treatment, whereas fungal taxa shown exhibit significant changes as determined by Welch’s t-test (P<0.05) and prevalence greater (>5%).
A comparison of the microbiota composition of subjects at baseline compared to PP alone highlights the effect of PP alone (Table 5). Notably, Prevotella nanceiensis was 0.00365%, at baseline whereas in PP alone, it decreased to 0.00014%. This corresponds to a fold change of 21.4, (P=0.02) suggesting a substantial reduction in the relative abundance of P. nanceiensis following pea protein supplementation alone.
|
Taxa |
|
Baseline RA (%) |
PP alone RA (%) |
Baseline Prevalence (%) |
PP alone Prevalence (%) |
Baseline: PP alone (FC) |
Elavated in |
Baseline: PP alone (p value) |
|
Bacterial Phyla |
Gemmatimonadetes |
0.0126 |
0.0053 |
68.4 |
65.6 |
2.0 |
Baseline |
0.018 |
|
Bacterial Genus |
Roseburia |
1.03 |
0.72 |
97.4 |
96.9 |
2.1 |
Baseline |
0.039 |
|
Achromobacter |
0.29 |
0.70 |
71.1 |
78.1 |
-15.0 |
PP alone |
0.017 |
|
|
Parvimonas |
0.0031 |
0.0022 |
23.7 |
28.1 |
-4.1 |
PP alone |
0.021 |
|
|
Cellulosimicrobium |
0.0276 |
0.0033 |
31.6 |
25.0 |
-9.3 |
PP alone |
0.040 |
|
|
Salinispora |
0.0024 |
0.0010 |
23.7 |
18.8 |
-8.4 |
PP alone |
0.024 |
|
|
Prevotella nanceiensis |
0.00365 |
0.00014 |
21.1 |
3.1 |
21.4 |
Baseline |
0.020 |
|
|
Fungal Genus |
Trametes |
0.296 |
0.061 |
44.7 |
28.1 |
4.8 |
Baseline |
0.033 |
|
Fungal Species |
Candida flosculorum |
0.00016 |
0.02640 |
7.9 |
15.6 |
-165.6 |
PP alone |
0.033 |
Table 5 Changes in Bacteriome and Mycobiome Composition Comparing Baseline to PP alone
A comparison of the bacteriome composition of subjects treated with PP+BFX compared to PP alone (Table 6) provides insights into the response to BFX supplementation when pea protein is included in both groups. The phyla Bacteroidetes exhibited a higher relative abundance in PP+BFX (35.1%) compared to PP alone (26.9%), with a fold change of 1.3 (P=0.034). Similarly, Spirochaetes and Cyanobacteria displayed increased relative abundances in PP + BFX, with fold changes of 1.6 (P=0.039) and 7.5 (P=0.049), respectively.
|
Taxa |
PP+BFX RA (%) |
PP alone RA (%) |
PP+BFX Prevalence (%) |
PP Alone Prevalence (%) |
PP+BFX: PP alone (FC) |
Elavated in |
PP+BFX:PP alone (p value) |
|
|
Bacterial Phyla |
Bacteroidetes |
35.1 |
26.9 |
100 |
100 |
1.3 |
PP+BFX |
0.034 |
|
Spirochaetes |
0.028 |
0.018 |
89.5 |
78.1 |
1.6 |
PP+BFX |
0.039 |
|
|
Cyanobacteria |
0.074 |
0.010 |
92.1 |
75.0 |
7.5 |
PP+BFX |
0.049 |
|
|
Bacterial Genus |
Bilophila |
0.382 |
0.197 |
89.5 |
87.5 |
1.9 |
PP+BFX |
0.027 |
|
Leadbetterella |
0.001035 |
0.00021 |
26.3 |
9.4 |
4.9 |
PP+BFX |
0.040 |
|
|
Pyramidobacter |
0.00105 |
0.00015 |
23.7 |
3.1 |
6.9 |
PP+BFX |
0.045 |
|
|
Bacterial Species |
Eubacterium dolichum |
0.172 |
0.456 |
81.6 |
87.5 |
-2.6 |
PP alone |
0.016 |
|
Bulleidia moorei |
0.0051 |
0.0155 |
31.6 |
34.4 |
-3.1 |
PP alone |
0.041 |
|
|
Corynebacterium simulans |
0.032 |
0.197 |
36.8 |
53.1 |
-6.2 |
PP alone |
0.050 |
|
|
Fungal Genus |
Brodoa |
0.000245 |
0.002055 |
10.5 |
18.8 |
-8.4 |
PP alone |
0.031 |
|
Fungal Species |
Candida khao-thaluensis |
0.000412 |
0.005699 |
7.9 |
15.6 |
-13.8 |
PP alone |
0.037 |
|
Sclerotinia homoeocarpa |
0.0105 |
0.001326 |
28.9 |
12.5 |
7.9 |
PP+BFX |
0.042 |
Table 6 Changes in Bacteriome and Mycobiome Composition Comparing PP+BFX to PP alone
Bacteroides uniformis was highest in PP+BFX when compared to the baseline and PP alone, whereas the abundance of Saccharomyces cerevisiae was higher in PP+BFX compared with PP alone, as shown in Supplemental Figure 1. The mycobiome composition was more diverse in respects to the PP+BFX treatment with the key finding that Galactomyces geotrichum was significantly elevated when compared to baseline (P=0.046) (Table 4). G. geotrichum had a relative abundance of 4.1% at baseline and 12.1% in PP+BFX, demonstrating a 2.9-fold increase. Although not statistically significant, Candida albicans (39.7%) was decreased in the PP+BRF treatment compared to baseline (47.4%) or PP alone (49.8%) (Supplemental Figure 2).
The pathogens Escherichia coli, Prevotella copri, Shigella flexneri, and Brevundimonas diminuta were decreased in PP+BFX treatment when compared to PP alone, as shown in Supplemental Figures 3A & B.
The primary aim of the current study was to determine if addition of the probiotic, BIOHM FX (BFX), to a pea protein supplement (15g pea protein-PP) altered the gut microbiome balance and enhanced the absorption of non-animal proteins determined via quantification of essential amino acids (EAAs) in plasma. A placebo-controlled crossover clinical study in active men (n=39 finishers) was performed and stool samples were analyzed for changes in microbiome composition from baseline and compared for PP versus PP+BFX. Plasma EAAs were measured pre- and post-ingestion of the pea protein +/-BFX for up to 180 min. Self-reported changes in gastrointestinal (e.g., bloating, flatulence) and quality of life (e.g., fatigue, mood, and energy) indices were also measured. We observed that subjects ingesting PP+BFX exhibited a distinct microbiome profile compared to subjects at baseline and following ingestion of PP alone. Differences were also observed in plasma EAA at 120 min post-ingestion, wherein PP+BFX values were greater than PP. Microbiome analysis of stool samples demonstrated that bacterial pathogens decreased following ingestion of PP+BFX compared to PP. Fungal species in the gut were also altered following consumption of PP+BFX. Interestingly, the abundance of several Pseudomonas species, cyanobacteria phyla and the fungal species Galactomyces geotrichum was elevated when the combination of PP+BFX were consumed by study subjects.
Previous investigators have shown that individuals consuming probiotic supplements containing lactic acid producing bacteria or Bifidobacterium have improved mucosal layer integrity and increased nutrient absorption.23,24 Early studies of biofilm formation have shown that polymicrobial communities form within polysaccharide-rich extracellular matrices, with negative consequences that include alteration of gut permeability, decreased antimicrobial susceptibility, and reduction of host immune response.25 In this regard, for proteins to be maximally absorbed, they must first be broken down into amino acids or oligopeptides by enzymes from enterocytes within the mucin membrane of the small intestine8,26 with the expression of these enzymes in direct correlation to mucin thickness.9 However, if partially digested food from the stomach does not come into direct contact with the mucosal layer, absorption cannot occur. In recent in vitro biofilm studies, the permeability of casein through an epithelial cell monolayer was significantly increased (P=0.0001) by the addition of BFX in the presence and/or absence of mixed-species biofilms.13 Thus, these data provide evidence that BFX is capable of increasing the permeability of the epithelial lining of the gut.
In a similar study, investigators showed that plasma levels of EAAs tryptophan, and cysteine, and total amino acids were increased in a cohort of older women consuming plant-derived protein supplemented with the probiotic Weizmannia coagulans GBI-30, 6086 (BC30, formerly classified as Bacillus coagulans), at a level (4.4% increase) very similar to the observed change reported here for EAA.27 Further, in a recent comparable clinical study, probiotic administration with pea-derived protein significantly increased methionine, histidine, valine, leucine, isoleucine, tyrosine, total BCAA, and total EAA maximum concentrations as compared to pea protein alone.4 In this study, based on in vitro data, the investigators hypothesize that increased proteolysis and a synergistic effect with the combination of two probiotic strains (L. paracasei LP-DG® and L. paracasei LPC-S01) led to increased EAA absorption.
Interestingly, in this study, Pseudomonas species were elevated when the combination of PP+BFX were consumed by the study subjects. This observation is in agreement with a previous study showing Pseudomonas species following probiotic consumption.28 Pseudomonas species are known to be involved in protein metabolism via their ability to degrade and utilize proteins as a source of nutrients. Although often associated with the pathogen Pseudomonas aeruginosa, many more Pseudomonas species are known, and the majority are not pathogenic, indeed beneficial Pseudomonas species are also known.29 These bacteria possess a range of proteolytic enzymes, such as proteases, peptidases, and exopeptidases, which enable them to break down complex proteins into smaller peptides and amino acids. This protein degradation capacity allows Pseudomonas to access nitrogen and carbon sources necessary for their growth and survival.30–32 Therefore, it is interesting to hypothesize that one of the potential underlying mechanisms by which BFX incorporation to PP increased amino acid absorption is by increasing the abundance of Pseudomonas spp. that enhanced pea protein degradation via endogenous increases in proteolytic enzymes, thus enhancing EAA detected in the plasma.
Table 6 provides evidence that the combination of PP+BFX supplementation has a significant impact on the gut microbiome. The PP+BFX treatment demonstrated an increase in the abundance of beneficial bacterial species, highlighting a potential synergistic effect between pea protein and BFX. In contrast, PP alone did not exhibit as many significant changes in the gut microbiome profile (Table 5).
An interesting key effect of BFX supplementation is an increase in the abundance of Bacteroidetes and Cyanobacteria when supplemented with PP+BFX compared to PP alone (Table 6). Bacterial species belonging to these phyla, contribute to the maintenance of gut homeostasis, nutrient metabolism, immune regulation, and overall host health.33 Furthermore, Cyanobacteria has been shown to help in fiber (prebiotics) breakdown, and to produce a-amylase.34,35 In addition, the breakdown of dietary fibers by anaerobic intestinal microbiota has been previously reported to produce short-chain fatty acids (SCFAs) which have been reported to exert multiple beneficial effects on mammalian energy metabolism.36–38
In the current study, BFX was shown to positively modulate the gut microbiome by influencing the abundance of several species. Importantly, the abundance of S. cerevisiae and Galactomyces geotrichum in stool samples following BFX ingestion increased dramatically from baseline and was much higher than the placebo group (PP alone). In a review of the gut mycobiota, Wu et al. noted that benefits of healthy levels of S. cerevisiae include the capacity to lessen the severity of gastroenteritis, prevent adherence of adherent-invasive E. coli, (which colonize the ileal mucosa of CD patients) in a mouse model, and relieve abdominal pain in irritable bowel diseases.39 Additionally, G. geotrichum, a commensal fungus, is recognized for its ability to produce vitamin B2 and peptides that inhibit the angiotensin I converting enzyme,40,41 thereby suggesting a potential role in maintaining gut health.
The abundance of the fungal pathogen C. albicans was lower in participants ingesting the PP+BFX-fortified supplement compared to baseline and the PP alone group (Supplemental Figure 1). C. albicans is a normal commensal fungal strain in the gut, but much attention has recently been given to multi-symptom conditions caused by C. albicans overgrowth. This overgrowth may be attributed to an imbalanced diet high in sugar and other refined carbohydrates that can cause a variety of conditions such as constipation, diarrhea, nausea, gas, cramps, and bloating.42,43 Within the bacterial species isolated, abundance of the probable pathogens E. coli, P. copri, S. flexneri, and B. diminuta was lower under the PP+BFX treatment than the PP alone treatment.
While PP alone can implement change, the combination of PP+BFX results in distinct alterations in the microbiota, enhancing the relative abundance of certain beneficial microbes and reducing others. This highlights the potential for dietary interventions such as PP+BFX to modulate the gut microbiome, which may have important implications for health given the microbiome's role in many aspects of human physiology, including digestion, immune function, and even mood regulation.
One of the major limitations of this study was the crossover design, which although implemented to reduce bias in the protein absorption assay and increase statistical power by using each subject as their own control, had a potential impact on the microbiome analysis. This can be observed in Supplemental Figure 1, where the relative abundance of S. cerevisiae was low at baseline and increased substantially in PP+BFX but was lower in PP alone. Due to BFX containing S. boulardii (a sub-species of S. cerevisiae), this suggests increased abundance in the PP+BFX treatment was due to consumption of the BFX. However, in PP alone treatment, which had smaller amounts of this fungi, the lower abundance may be explained by the prior exposure of half of the subjects in the PP treatment had already been exposed to S boulardii in the first 4weeks of treatment prior to crossing over to the PP alone group. A second limitation was the lack of gathering information on baseline GI health, as only acute GI markers (gas and bloating on visits 3 and 4) were evaluated.
These findings underscore the importance of considering the combined effects of dietary interventions on the gut microbiome and suggest that the simultaneous supplementation of pea protein and BFX may have a more profound influence on gut microbial composition. Further research is warranted to elucidate the underlying mechanisms and explore the clinical implications of these findings.
In summary, our study indicated that the addition of BFX to the pea protein supplement resulted in a distinct change in the microbiome profile compared to baseline and ingestion of PP alone. Participants ingesting PP+ BFX had a significantly higher plasma EAA concentration (P= 0.047) at 120 min post-consumption as compared to PP alone. While several other commercially available probiotics have been shown to increase amino acid absorption, BFX appears to enhance the gut microbiome balance of organisms capable of producing increased levels of proteolytic enzymes, possibly leading to increased protein absorption (e.g., Psuedomonas spp.). Larger studies are warranted to confirm these findings and determine if there are additional benefits of BFX as an additive to plant protein supplements.
Conceptualization, Mahmoud Ghannoum; Data curation, Michael La Monica, Tim Ziegenfuss and Mauricio Retuerto; Formal analysis, Michael La Monica, Mauricio Retuerto and Thomas McCormick; Funding acquisition, Mahmoud Ghannoum; Investigation, Michael La Monica, Tim Ziegenfuss, Betsy Raub, and Mauricio Retuerto; Methodology, Mauricio Retuerto, Michael La Monica, and Thomas McCormick; Project administration, Tim Ziegenfuss, and Mahmoud Ghannoum; Writing – original draft, Michael La Monica and Tim Ziegenfuss; Writing – review & editing, Michael La Monica, Mauricio Retuerto and Thomas McCormick.
This work was supported in part by a grant from BIOHM Health, LLC.
Informed consent was obtained from all subjects involved in the study.
All data generated or analyzed during this study are included in this published article (and its supplementary information files). The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
We would like to thank the dedicated group of subjects who participated in this study. The presentation of results of this study does not constitute endorsement by any of the researchers or their affiliations.
This research was sponsored in part by a grant from BIOHM Health, LLC. The presentation of results of this study does not constitute endorsement by any of the researchers or their affiliations. BIOHM Health, LLC had no role in the collection, analyses, or interpretation of the data. Mahmoud Ghannoum is the founding partner of BIOHM Health, LLC. Michael La Monica, Tim Ziegenfuss, and Betsy Raub are employees of The Center for Applied Health Sciences.

©2023 Retuerto, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.