eISSN: 2373-6372


Research Article Volume 10 Issue 3
1Department of Internal Medicine, Cairo University, Egypt
2Department of Pathology, Cairo University, Egypt
3Department of Biochemistry Cairo University, Egypt
4Department of Clinical and Chemical pathology, Cairo University, Egypt
Correspondence: Serag Esmat, Professor, Department of Internal Medicine, Gastroenterology Division, Faculty of Medicine, Cairo University, Egypt
Received: February 19, 2019 | Published: June 7, 2019
Citation: Naga M, Amin M, Esmat S, et al. Hemochromatosis genes and HCV in Egyptian patients; is there a relationship? Gastroenterol Hepatol Open Access. 2019;10(3):145-152 DOI: 10.15406/ghoa.2019.10.00373
Aim: To evaluate the prevalence of HFE mutations in Egyptian patients with chronic HCV and its relation to the response to treatment.
Methods: 657 Chronic HCV patients and 160 healthy age and sex matched control group were studied. The frequency of the genotypes among the patients and the control group were performed. The 657 patients were candidates for pegylated-interferon/Ribavirin treatment. According to the response of treatment we divided the patients into two groups; responders (n=356) and non-responders (n=301).
Results: There was a statistically significant difference in the frequency of HFE 845G>A, HFE187C>G and HFE193A>T genotypes between responders, non-responders and controls (P<0.001) . There was a statistically significant association between G allele of HFE 845 G>A, the C allele of HFE187C>G, A allele of HFE 193A>T and response to interferon. Carriers of the A allele of HFE 845 G>A had O.R.10.3 (95%C.I. 7.1-14.8). Carriers of G allele of HFE187C>G had O.R. 9.3 (95%C.I. 7.2–11.9), while carriers of T allele of HFE193A>T gene mutation had O.R. 9.2 with 95% C.I. 7.2–11.8.
Conclusion: HFE gene polymorphism in chronic HCV Egyptian patients may play a role in the disease progression and can affect the response to treatment.
Keywords: Hemochromatosis gene mutations, liver fibrosis, iron deposition, HCV treatment response; chronic HCV
Hepatitis C virus (HCV) is a main worldwide clinical troublesome and considered the main reason of chronic liver disease, cirrhosis and hepatocellular carcinoma.1–3 It has been estimated that around 170million people among the whole world population have HCV.4 Egypt is considered to be the highest country in HCV prevalence worldwide, affecting around of 10%-20% of the people in rural regions5 and more than 500,000 newly infected per year as mentioned in the Egyptian Demographic Health Survey in 2009.6 Numerous research efforts have been made to treat HCV specially in epidemic countries.7 The high cost and adverse events of the therapy and need for close follow up essentially made a burden on healthcare in Egypt resulting in a reduction of services provided to HCV patients. Thus, the availability of dependable predictors to viral response is inevitable for appropriate selection of patients who are candidates for treatment.
Several genetic factors may have an effect on the virological response to antiviral therapy which may explain the variation in the response rates among individuals.8,9 Hereditary hemochromatosis (HH) is an inherited autosomal recessive condition of the metabolism of iron. It results from increased intestinal absorption of iron leading to its accumulation in hepatocytes, pancreatic cells, cardiac cells and other organs leading to destruction to their construction and decline of their function.10 In 1996 the responsible gene HFE was discovered and some HFE mutations were described. In Europe most of the cases showed homozygous 845G→ A mutation in exon 4 of the HFE gene, this led to change in amino acid at position 282 to tyrosine instead of cysteine: C282Y.10,11 The mutant allele 187C→ G was found to be more commonly appears in the exon 2 of the HFE gene where aspartate substitutes histidine at position 63 (H63D).11 The HFE mutations are considered the leading gene disparities in hereditary hemochromatosis with iron burden in Caucasians. High hepatic iron concentration was frequently present in chronic hepatitis patients12 and may intensify the oxidative load, and fasten the development of fibrosis.13
The mutations of HFE was mentioned to be related with favorable response to interferon14,15 most studies on HFE mutations was in Western countries, and few studies has been conducted in an Asian countries.16 HH expresses ethnicity differences and the information about the relation of HH mutations in Egyptian patients with chronic HCV is lacking. Therefore we aimed in our study to evaluate the effect of HFE mutations in HCV Egyptian patients.
Six hundred and fifty seven HCV patients were included in the study and received pegylated-interferon/ ribavirin therapy. Patients were enrolled from the medical clinics of, Kasr Al Aini hospital, National liver institute and Beni Suef community hospital. One hundred and sixty healthy subjects that matched the age and sex were included as a control group.
Patients and controls were subjected to complete physical examination including measurement of the body mass index and laboratory investigations: total bilirubin, direct bilirubin, serum alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, creatinine, complete blood count, fasting blood sugar, HbA 1c, albumin, prothrombin time and concentration, INR, hepatitis B surface antigen HCV antibody, total Iron binding capacity, ferritin & iron levels, transferrin, transferrin saturation, soluble transferrin receptor (sTFR), and hemochromatosis genotype studying.
For the HCV patients the following was done; quantitative polymerase chain reaction for detection of hepatitis C virus RNA, thyroid stimulating hormone, antinuclear antibodies, abdominal ultrasound and liver biopsy. At the time of the start of the study the newly developed direct anti-viral inhibitors were not yet established to treat of HCV in Egypt. The HCV patients received antiviral therapy of pegylated -interferon once weekly with 10.6 mg/kg ribavirin orally daily. Patients with a negative PCR or with more than two logs reduction after twelve weeks treatment were considered responders and continued treatment for 48 weeks. While those with less than two logs reduction in PCR levels after twelve weeks therapy were considered as non-responders. According to the response of treatment patients were into two groups: responders (356) and non-responders (301).PCR testing was repeated for responders six months after the end of treatment after to check the sustained viral response (SVR).
Types of interferon used
Pegylated interferon alfa-2a (PEGASYS® 180mcg/week), Pegylated alfa -2a (Reiferon Retard®160mcg/week) and Pegylated alfa-2b(PEGINTRON® 1.5mcg/kg/week) were used depending on their availability.
Iron status
Iron status assessment for each patient was assessed using biochemical tests including estimation of iron (µg/dl) and TIBC (µg/dl) using colorimetric kit (Stanbio Labrotary, Boerne, TX USA), ferritin (ng/ml) using ELISA (Diagnostic automation, INC cat#1601Z), transferrin (mg/dl) using ELISA (Diagnostic automation, INC) and soluble transferrin receptor (sTFR) (nmol/L) was detected using (DRG International Inc., Springfield, New Jersey, USA) ELISA kit. Transferrin saturation was measured from TIBC & iron and.
Hemochromatosis genotyping
For HEF genotyping allelic discrimination we applied TaqMan Analysis. All DNA samples for patients and controls were genotyped at HFE_187_C > G (His63Asp, rs1799945) , HFE845 G > A (Cys282Tyr, rs1800562) and HFE_193_A > T (Ser65Cys, rs1800730). Real time Polymerase chain reactions were done in reaction volumes of 25 µL containing 50 ng DNA, 10µl TaqMan Universal Polymerase chain reactions Mix with the passive reference ROX (Perkin Elmer), 280nmol/L of each primer, 6-carboxyfluorescein marked minor groove probe, and VIC-marked minor groove probe (400nmol/L for HFE_187_C > G and 200nmol/L for HFE_193_A > T). Table 1 shows the sequences of the Primer as well as the minor groove binder probes.
HEF primers |
HFE187C >G |
F 5'-GAAGCTTTGGGCTACGTGGAT-3' |
HFE193A>T |
F 5'-GAAGCTTTGGGCTACGTGGAT-3' |
|
HFE845G>A |
F 5'-GGCTGGATAACCTTGGCTGTAC-3' |
Table 1 Primers and probes used for the Hemochromatosis gene
Liver biopsy was done after 6 hours fasting under complete aseptic precautions and local anesthesia with Tru-Cut biopsy needles size18 gauges. Biopsies were performed guided by ultrasound machine with convex probe, 3.5 MHz of GE LOGIQ® P5.The biopsies were saved in 10% formaldehyde then sent for histopathological examination in the same day. After the procedure, patients were monitored for 2-4 hours.
Histopathological examination of liver biopsies: at 5 microns 3 paraffin wax sections were cut and stained by Hematoxylin and Eosin stain for histopathological analysis and scoring the necro-inflammatory stage. For the degree of fibrosis Masson Trichrome stain was used. Prussian blue was used to check the presence of iron that appears as blue granules within the cytoplasm. Both modified HAI and Metavir scores were used for necro-inflammatory scoring and staging.
The study protocol was approved by the review board and ethical committee of the department of Internal Medicine, Faculty of Medicine, and Cairo University according to the Declaration of Helsinki.
Statistical analysis
Data was and entered after coding by the software SPSS (Statistical package for social science) version 21. Percentage and frequency were applied for qualitative variables. Mean with standard deviation was applied for parametric data. Percentiles & medians were used for quantitative variables. Kruskal Wallis test & ANOVA test were used to compare groups. Fischer test and Chi square test were applied for qualitative variable. The Mann Whitney test & t test were applied for comparison between two groups. The odds ratio with their 95% confidence intervals (OR, CI) was measured to assess the degree of association between each genotype & allele in patients and controls. The P-value was considered significant when less than 0.05. Testing deviation from the Hardy-Weinberg Equilibrium is usually performed by .
Table 2 shows the biochemical demographic characteristics of the studied groups.
|
Cases |
Control |
P value |
Age, mean±SD |
42.5±10 |
36.1±12.7 |
<0.001 |
Sex, no (%)** |
399(60.7) |
122(76.2) |
<0.001 |
AST, U/L* |
37(25-59) |
27(23-31.7) |
<0.001 |
ALT, U/L* |
40(23 -63) |
28(23-32) |
<0.001 |
Albumin, g/dl |
4.1±1.5 |
4.2±0.5 |
0.591 |
S.iron, ug/dl |
43.4±10 |
131.3± 31.9 |
<0.001 |
TIBC, ug/dl |
210.7±60 |
260± 42.1 |
<0.001 |
Ferritin, ng/ml |
188±65 |
190.9±38.8 |
0.463 |
Transferrin saturation% |
22.3±8.3 |
51.3±14.8 |
<0.001 |
s.Transferrin receptor, nmol/L* |
14.7(13.2 -23.5) |
11.5(10.5-12.3) |
<0.001 |
Transferrin, mg/dl* |
203(168-270) |
295(244-352.75) |
<0.001 |
Table 2 Demographic and biochemical characteristics of the patients and controls
The data are mean± S.D. unless otherwise indicated *Median (25th-75th percentile).
**no (%)
Biochemical results and HAI score in the 2 groups (responders and non-responders)
The median level of AST, ALT and the mean level of TIBC, BMI and HAI score were found to be significantly higher in the non-responders group versus responders. The mean levels of alkaline phosphatase, ferritin and TLC and median level of s. transferrin receptor were higher significantly in responders versus non responders group. There was no statistical significant difference in the ANA result in both groups of cases (Table 3).
|
Non-responders |
Responders |
P value |
Total Bilirubin, mg/dl * |
0.8(0.59-0.9) |
0.8(0.68-0.90) |
0.5 |
AST, U/L* |
52(36-72) |
28.5(19.3-43.7) |
<0.001 |
ALT, U/L* |
57(37-80.5) |
27(18-44) |
<0.001 |
Albumin, g/dl |
4.1±0.4 |
4.1±2.0 |
0.817 |
Alkaline phosphatase, U/L |
114.5±60.2 |
147.9±52.6 |
<0.001 |
S.iron, ug/dl |
43.1±10.0 |
43.5±10.1 |
0.589 |
TIBC, ug/dl |
228.5±56.8 |
195.6±59.4 |
<0.001 |
Ferritin, ng/ml |
158.4±39.3 |
212.9±72.4 |
<0.001 |
Transferrin saturation% |
20.0±6.6 a |
24.3±9b |
<0.001 |
s.transferrin receptor, nmol/l* |
14.5(13.5 -25.2) |
14.8(13.2-20.5) |
0.003 |
Transferrin, mg/dl * |
205(155-290) |
202.5(171-244) |
0.198 |
PCR before* |
320614(70534-1067500) |
213000(56462-648750) |
0.552 |
TSH* |
1.3(0.9-2.05) |
1.3(0.9-2.1) |
0.665 |
Hb |
13.7±1.5 |
13.9±1.8 |
0.09 |
TLC |
5936.7±1669.9 |
6491±2750 |
0.002 |
platelets |
206887±64201 |
200949±64307 |
0.238 |
PC% |
87.3± 9.9 |
88.1±8.8 |
0.26 |
PT |
12.7±0.6 |
13.09±6.6 |
0.380 |
INR |
1.09±0.09 |
1.09±0.09 |
0.505 |
ANA** |
300(99.7%) |
353(99.2%) |
0.629 |
BMI, kg/m2 |
28.8±4.7 |
26.3±4.2 |
<0.001 |
HAI score |
7.2±3.0 |
6.3±2.7 |
<0.001 |
Table 3 A comparisons of the biochemical results and HAI score between the responders and non-responders
The data are mean±S.D. unless otherwise indicated.*Median (25th-75th percentile).
**no (%)
The impact of the type of interferon used
No statistical significant difference was found regarding the type of interferon used between responders and non-responders P value 0.164 (Figure 1).
Genotypes of gene polymorphisms frequencies
All gene polymorphisms (HFE 187 C>G, HFE193A>T and HFE845G>A) were consistent with HWE (P values; 0 .082, 0.095 and 0,636).Table 4 shows theGenotypes of gene polymorphisms frequencies of the studied groups. A statistical significant difference was found in the frequency of genotypes of all polymorphisms studied among groups.
|
Non responders, n= 301 |
Responders, n=356 |
CONTROL, n= 160 |
P value |
HFE 187 C>G* |
35(11.7) |
240(67.4) |
101(63.1) |
<0.001 |
HFE 193A>T* |
47(15.6) |
242(68) |
94(58.8) |
<0.001 |
HFE 845 G>A |
97(32.2) |
318(89.3) |
145(90.6) |
<0.001 |
Table 4 Frequency of the genotypes of gene polymorphisms in the studied groups
All data are no (%)
Effect of allele type and response to therapy
The relation between the allele frequency and response to therapy are presented in table 5. A statistical significant association was present between the C allele of HFE 187 C>G gene polymorphism and response to treatment with interferon in patients with HCV. The carriers of T allele of HFE 193 A>T gene polymorphism are 9.2 times risk for not responding to interferon. Carriers of the A allele of HFE 845 G>A were 10.3 times risky for non-responding to interferon t than those of G allele. The carriers of G allele were 9.3 times risky for non-responding to interferon.
HFE 187 C>G* |
Non responders, n= 301 |
Responders, n=356 |
P value |
O.R(95%CI) |
G |
438(72.8) |
159(22.3) |
<0.001 |
9.3(7.2-11.9) |
C |
164(27.2) |
553(77.7) |
|
1 |
|
|
|
|
|
HFE 193 A>T* |
|
|
|
|
T |
429(71.3) |
151(21.2) |
<0.001 |
9.2(7.2-11.8) |
A |
173(28.7) |
561(78.8) |
|
1 |
HFE 845G>A |
|
|
|
|
A |
221(36.7) |
38(5.3) |
<0.001 |
10.3(7.1-14.8) |
G |
381(63.3) |
674(94.7) |
|
1 |
Table 5 Allele frequency and odds ratio in non-responders versus responders
All data are no (%)
Impact of hepatic iron deposition and response to treatment
No statistical significant difference was found between responders and non-responders regarding the deposition of hepatic iron P value 0.94 (Figure 2). When we studied the relationship between HFE gene polymorphisms and Prussian blue results we found no significant association between HFE 187 C>G, HFE 193 A>T or HFE 845G>A genotypes and Prussian blue results. The association of the different HEF genes studied with Metavir scoring system for activity and fibrosis are shown in Figure 3.
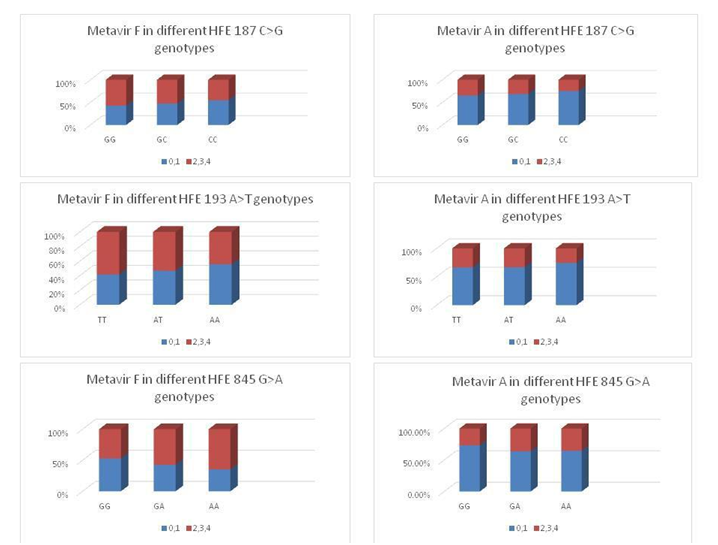
Figure 3The association of the different HEF genes studied with Metavir scoring system for activity and fibrosis.
The most common side effects that appeared in approximately 25% of the patients were:
Flu-like symptoms, loss of appetite, weight loss, insomnia, temporary hair loss, mild anemia, and mild leucopenia. However these side effects were mild and did not necessitate stopping the treatment and all the HCV patients finished the treatment.
657 Chronic HCV Egyptian patients were included in this study who received pegylated-interferon & ribavirin treatment and 160 healthy control subjects. Responders to treatment were 356 patients and 301 patients were non responders. The response rate to treatment was 54.18 % which is close to the response rate reported in several studies on Egyptian patients with Chronic HCV.17–21
The information about relationship between the HFE gene mutations and viral response to IFN & Ribavirin treatment was debatable.22–25 An association between H63D HFE polymorphism and higher viral response rates was showed in some studies.26This association may be explained by that the HFE gene is present on the short arm of chromosome 6, neighboring the major histocompatibility complex. The major histocompatibility complex genes translate the human leukocyte antigens, and are vital in antigen regulation and exhibition of T cells and CD8 +, CD4.27 some studies reported the impact of major histocompatibility complex gene variants and response to treatment in HCV infection.28–30 The results of our study contradict the described relationship between H63D mutation and higher virological response rate.28 In our study carriers of the H63D mutation specially carriers of the G allele (HFE 187 C>G) were 9.3 times risky for non-responding to interferon.
The clarification of the negative effect of H63D mutation on the virological response seen in our study may be correlated to the characteristics of the ethnicity of our patients because all the patients in our study were Egyptians with HCV genotype 4. On the other hand the results of this study are similar to the study of Coelho-Borges et.al that included Brazilian patients with HCV genotypes other than genotype-1 who mentioned the negative effect of H63D mutation on the sustained virological response to IFN & Ribavirin treatment and furthermore they concluded that the finding of HFE polymorphism is a durable predictor of non-response in his study.31
The results of our study also showed that Carriers of S65C mutations specifically the T allele (HFE 193 A>T) gene polymorphism have 9.2 times risk for not responding to interferon than non-carriers which can be considered a predictor of response to Interferon based therapy. In 2012 Ishizu et.al studied the effect of HFE mutations in Japanese patients with HCV; their study included 251 CHCV patients. Their results showed that 5.6% of the patients had the H63D mutation and not S65C alterations. More over all the fourteen patients with H63D mutations responded well to the IFN/Ribavirin therapy.32 Another study in 2012 by Li SHet.al performed a meta-analysis which included seven studies from seven different countries ; Norway, Poland, Portugal, France, China, Brazil, and USA that included 1884 chronic HCV patients in order to assess the effect of HFE mutations on the virological response to interferon-based antiviral therapy. They concluded that H63D alteration in HFE is linked with a greater sustained viral response rate in HCV patients who received interferon treatment while S65C mutations had a very low frequency which prevented them to evaluate its effect on viral response.33
On the other hand the carriers of the A allele of HFE 845 G>A showed a 10.3 odd ratio of no response to interferon than those of G allele which was the highest risk of non-response to treatment among all HFE polymorphisms in our study.
In the current study the mean levels of ferritin and median level of s. transferrin receptor were greater in responders versus non responders. There statistical insignificant difference among responders and non-responders regarding the hepatic iron deposition.
More over when we studied the relationship between HFE gene polymorphisms and Prussian blue results we found no significant association between HFE 193 A>T, HFE 187 C>G, or HFE 845G>A genotypes and Prussian blue results.
However, the increased risk of non- responding to IFN/RBV treatment in the patients with these HFE genetic polymorphisms in our study was possibly indirectly linked to the overload of iron. These findings are supported with the fact that many studies, couldn’t find a significant dissimilarity in hepatic iron deposition between HCV responders and non-responders patients after they received the IFN/RBV treatment.34–37
The elevated ferritin levels in HCV patients may be attributed to the fact that ferritin is considered as acute phase reactant which may act as an indicator of more active liver disease and it has been reported that chronic HCV patients who have elevated serum ferritin also showed more liver fibrosis and inflammation when compared to patients with average ferritin levels.24,38,39
The association of HFE polymorphisms specially the carriers of the A allele of HFE 845 G>A, carriers of C allele of HFE 187 C>G gene and the T allele of HFE 193 A>T gene polymorphisms with the highly increased risk of non-responding to IFN/RBV can be well thought-out as a strong predictor of non-response to IFN/RBV treatment in Egyptians infected with HCV. Identification HFE gene polymorphism in patients chronically infected with HCV might be considered as a valuable tool in selecting patients for treatment as it seems to be a factor in the response to treatment.
This work was funded by the Science and Technology Development Fund (STDF), Egypt, Grant No. 1587.
We appreciate the efforts of Professor Nouman Elgarim (Professor of internal medicine, Cairo University) in enable the recruitment of patients from Beni Suef public hospital. We would like to thank Dr.Ismail Kasem (Assistant Professor of internal medicine, National liver institute, Cairo) for facilitating the recruitment of patients from the National liver institute. We are thankful to Dr Amr Abdelaziz (Assistant lecturer of internal medicine, Cairo University) for his efforts in sample collection from Beni Suef public hospital. Supported by the Science and Technology Development Fund, Ministry of Scientific Research Egypt, Project No. 1587; and Cairo University.
All authors declare that they don’t have conflict of interest in any kind.
The review board and the ethical committee of the Department of Internal Medicine, Faculty of Medicine, Cairo University approved the study protocol, which was performed according to the Declaration of Helsinki.
All study participants provided informed written consent prior to study enrolment.
The technical appendix, statistical code, and dataset are available from the corresponding author at seragesmat@kasralainy.edu.eg. No additional data are available.

©2019 Naga, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 World Hepatitis Day is held on July 28th, 2024 and the purpose of this day is to raise awareness of hepatitis and to monitor the availability of related vaccines and encourage action to reduce suffering and death from the infectious disease which causes hepatitis. Initial diagnose and precautions will help you to have a healthy liver and peaceful life. So, it is an opportunity for to all the researchers to motivate action and submit your interesting papers on raising the awareness to encourage people to get proper diagnose from this infectious disease and the submissions received till July 24th will be offered with a best discount of 30% in Gastroenterology & hepatology: Open Access Journal.
World Hepatitis Day is held on July 28th, 2024 and the purpose of this day is to raise awareness of hepatitis and to monitor the availability of related vaccines and encourage action to reduce suffering and death from the infectious disease which causes hepatitis. Initial diagnose and precautions will help you to have a healthy liver and peaceful life. So, it is an opportunity for to all the researchers to motivate action and submit your interesting papers on raising the awareness to encourage people to get proper diagnose from this infectious disease and the submissions received till July 24th will be offered with a best discount of 30% in Gastroenterology & hepatology: Open Access Journal.