eISSN: 2373-6372


Background and aim: Exonuclease1 (EXO1) is a member of the RAD2 nuclease family which is involved in mismatch repair (MMR) system and contributes to the maintenance of genomic stability, modulation of DNA recombination and cell cycle arrest mediation. K589E (rs1047840) as a potentially functional polymorphism in EXO1 gene may alter cancer risk by influencing its repair activity.
Method: We designed a case-control study consisting of 319 subjects with colorectal cancer (CRC) and 310 healthy controls to investigate the effect of K589E polymorphism on CRC susceptibility and clinicopathological features in an Iranian population. Genotype determination was performed by PCR-RFLP method.
Results: We provided the first evidence that there is not any significant association between EXO1 K589E alleles and genotypes and risk of CRC, even after adjustment for sex, age and smoking status (OR=1.033; 95% CI 0.814-1.312). Also analysis of clinicopathological characteristics and genotype distribution did not show any significant correlation.
Conclusion: Despite the well-known role of EXO1, our results revealed that EXO1 K589E polymorphism could not be a potential predisposing risk factor in genetic susceptibility to CRC, at least in the studied population. A large scale case-control study will need to be carried out to further validate this polymorphism as a non-contributor to the risk of CRC in Iranian population.
Keywords: Colorectal cancer; Exonuclease1; Single nucleotide polymorphism
CRC: Colorectal Cancer; EXO1: Exonuclease1; SNP: Single Nucleotide Polymorphism; MMR: Mismatch Repair; MSI: Microsatellite Instability; WHO: World Health Organization; FFPE: Formalin-Fixed Paraffin- Embedded; PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism; MSS: Microsatellite Stable; OR: Odds ratio; SPSS: Statistical Package for Social Sciences; CI: Confidence Intervals; TNM: Tumor Node Metastasis
Colorectal cancer (CRC) is the third most common cancer-related death throughout the world. According to the World Health Organization (WHO) data set, the incidence of CRC is rising in Asia with an incidence rate of one million cases and mortality of more than 500,000 per year.1,2 Previous studies show that the burden of CRC is increasing in Iran.3-5 Genetically, colorectal cancer is the result of accumulation of genetic alterations and destabilization of the genome that is a prerequisite of carcinogenesis. Genetic instability is commonly caused by a defect in the DNA repair system.6 DNA mismatch repair (MMR) is a highly conserved DNA repair system which recognizes mismatched bases spontaneously incorporated during DNA replication and recombination.7,8 Exonuclease1 (EXO1) gene is one of the interesting genes for the study of cancer susceptibility. The EXO1 gene (EXO1; MIM#606063) belongs to the MMR system. EXO1 gene is located at chromosome 1q42-q43 and consists of one untranslated exon followed by 13 coding exons. This gene is transcribed to yield a 3-kb mRNA and encodes a 846-amino acid protein. This protein is a member of RAD2 nuclease family. It has a role in DNA replication, repair and recombination.9-12 Wei et al.,13 found that increased rate of infertility in Exo1-null mice is due to meiotic defect. It also causes reduction in survival and increasing in susceptibility to the arising lymphoma. Bardwell et al.,14 reported that mice with missing exon 6 of Exo1 had defective mismatch repair. Exo1 has a critical role as both 5'-3' and 3'-5' nuclease activity.15 This protein can form a complex with MMR proteins such as MSH2 (Exo1-MutLα) and MLH1 (Exo1-MutSα) in both yeast and human cells and with MSH3 in human cells.11,16,17 MutSα and MutLα have an ATPase activity. ATP binding and hydrolytic activities of hEXO1 are necessary for efficient MMR.18-20 Microsatellite instability (MSI) is caused by a failure of the MMR system. Because of strong interaction between Exo1 and MMR proteins such as MLH1 and MSH2, MSI status may be associated by EXO1 genotypes.21 Genetic polymorphisms in DNA repair genes such as EXO1 are thought to modulate DNA repair capacity and consequently are suggested to be associated with colorectal cancer.22 Single-nucleotide polymorphism (SNP) is a DNA sequence variation and has a great potential application to determine association with susceptibility to several cancers such as oral, breast, prostate, gastric and colorectal cancer.23-31 These indicate that substitution in these SNPs may effect on genes' functions or expression levels. So, risky genotypes have an effect on cancer susceptibility.20 Wu et al.,32 reported 12 missense single nucleotide polymorphisms in the EXO1 gene including K589E in exon11.21-32 An exonic SNP doesn't necessarily cause a dysfunctional protein. However influences of SNPs on biochemical interaction between components of the MMR pathway were reported by recent studies.33,34 According to well-defined role of defective MMR in CRC, it is important to inspect if common variants in the involved genes affect susceptibility to the disease at a population level. Several studies have reported K589E as the genetic risk factor of oral, breast, lung and gastric cancers.17,35-38 Our recent knowledge shows that there isn't any report to evaluate K589E polymorphism and risk of CRC. Based on above knowledge, the aim of this study was to investigate a relationship of K589E polymorphism of the EXO1 gene with the risk of CRC and clinicopathological features.
Study population and sample collection
Three–hundred and nineteen CRC patients diagnosed with colorectal cancer were registered in cancer registry unit of the Gastroenterology and Liver Diseases Research Centre, Shahid Beheshti University of Medical Science, Tehran, Iran, from October 2007 to January 2011, with positive colonoscopy and pathology results for a colon or rectum malignant tumor. All patients voluntarily participated, completed a self-administrated questionnaire and provided peripheral blood samples. In all colorectal cancer patients, pathological grade and clinical stage were confirmed by operation and pathology. The histological classification and clinicopathological staging were according to the criteria of the UICC Tumor-Node-Metastasis classification of Malignant Tumors (TNM) 6th edition, 2002, colon and rectum (ICD-O C18-C 20).
The pathologic stage of colorectal tumors were classified into stage 0 (TisN0M0), stage I (T1-2, N0, M0), stage II (T3-4, N0, M0), stage III (any T, any N, M0), and stage IV (any T, any N, M1). Tumor grade was classified into high (poorly differentiated), intermediate (moderately differentiated), and low (well differentiated).5,39 Three hundred and ten of non-cancer volunteers were selected as controls at the same time. The control group included individuals without family history of gastrointestinal disorders and eligible colonoscopy report for malignancy, polyps or inflammatory ulcers. All procedures were approved by the ethics committee of the Gastroenterology and Liver Diseases Research Center.
DNA extraction
Peripheral blood and paraffin-embedded tissue extraction: A 5ml sample of peripheral blood from all subjects was collected into tubes including Ethylene Diamine Tetra Acetic Acid (EDTA) as anticoagulant and stored at 4ºC. Genomic DNA was purified from peripheral blood leukocytes using a phenol-chloroform standard protocol.40 Paraffin-embedded tissues were also available for 107 of patients. For these patients we extracted genomic DNA from tumor tissue in order to investigate correlation between Exo1 genotypes and MSI status. QIAamp DNA Mini Kit was used for DNA extraction from formalin-fixed, paraffin- embedded (FFPE) tissues.
Genotyping assays
Genotype determination was done by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) method. We utilized the sequences: 5'-AGATGTAGCACGTAATTCAACTG-3' as forward and 5'-AATCGCTCTTTCTTCGGAACTG-3' as reverse primer to detect their genotypes. Primer designing was done by Gene Runner software version 3.05 (Hastings Software Inc., USA) and NCBI database. NCBI Primer-BLAST was used to check primers for specificity. Amplification was performed using thermocycler (Mastercycler Eppendorf, Germany). In brief, PCR was performed in a 25μl PCR reaction mixture with approximately 100 ng genomic DNA, 0.5mM each dNTP, 1x PCR buffer, 0.4μM each primer, 3mM Mgcl2 and 1 U rTaq DNA polymerase (Takara, Kyoto, Japan). The amplification was performed by the following cycling conditions: for the SNP rs1047840 one cycle for initial denaturation at 95°C for 5min; 30 cycles of 95°C for 45s, 61°C for 40s, and 72°C for 45s; and a final extension at 72°C for 10min. The 257-bp PCR products were studied after digestion with MseI restriction enzyme (New England Biolabs, UK) at 37°C for 5h. MseI digestion gives 152 and 105-bp fragments for Lys allele and a single 257-bp fragment for Glu allele. To confirm genotyping results, approximately 10% of the samples were sequenced randomly with an automated genetic analyzer 3130xl (Applied Biosystem, USA).
Analysis of MSI
MSI analysis was performed for 107 patients whose tissues were available. Tumor and normal tissue DNA were analyzed for MSI using the most pentaplex assay version of five microsatellite markers: BAT-26, BAT-25, NR-21, NR-24 and NR-27 markers. Oligonucleotide forward primers were fluorescently 5'-labeled. Primer sequences are mentioned in Table 1. Amplification conditions were as follows: initial denaturation at 95ºC for 3min; 34 cycles of 95°C for 30s, 55°C for 30s, and 72°C for 30s, with a final extension of 5 min at 72°C. Then fragment analysis was performed by ABI genetic analyzer 3130xl (Applied Biosystem, USA) using Gene Mapper Analysis 4 software (Applied Biosystem). Tumors showing instability at least two of five markers were considered as MSI high (MSI-H). Tumors exhibiting MSI at only one or none of markers were defined as MSI low (MSI-L) and microsatellite stable (MSS), respectively.
Name |
Gene |
Gene Bank Number |
Repeat |
Primer Sequences (5' -> 3') |
Amplicon Size (bp) |
NR-27 |
Inhibitor of apoptosis protein-1 |
AF070674 |
27 A 5'UTR |
F: AACCATGCTTGCAAACCACT |
87 |
R: CGATAATACTAGCAATGACC |
|||||
NR-21 |
SLC7A8 |
XM 033393 |
21 T 5' UTR |
F: GAGTCGCTGGCACAGTTCTA |
109 |
R:CTGGTCACTCGCGTTTACAA |
|||||
NR-24 |
Zinc finger 2 |
X60152 |
24 T 3' UTR |
F: GCTGAATTTTACCTCCTGAC |
131 |
R: ATTGTGCCATTGCATTCCAA |
|||||
BAT-25 |
C-kit |
X06182 |
25 T intron16 |
F: TACCAGGTGGCAAAGGGCA |
153 |
R: TCTGCATTTTAACTATGGCTC |
|||||
BAT-26 |
hMSH2 |
U04045 |
26 A intron5 |
F: CTGCGGTAATCAAGTTTTTAG |
183 |
R: AACCATTCAACATTTTTAACCC |
Table 1 Oligonucleotide primers used for microsatellite analysis
Statistical analysis
Statistical Package for Social Sciences (SPSS) software version 13 (SPSS Inc., USA) was used to perform all statistical analyses in this study. Correlation between the distribution of the allele and genotype frequencies and also clinicopathological characteristics was examined using the χ2 test. To compare the observed genotype frequencies among studied controls and cases with the expected genotype frequencies, we used the χ2 test to evaluate Hardy-Weinberg equilibrium. Logistic regression analysis was used to adjust the data for probable confounding factors such as age, gender and smoking. Odds ratio (OR) are given with the respective 95% confidence intervals (95% CI). To calculate ORs and 95% CIs, the homozygous genotype for the Glu allele of EXO1 was used as the reference. The calculation of differences in quantitative and qualitative demographic variables was performed using Student's t-test or a χ2 test, respectively. Values of P<0.05 were considered significant.
The frequency distribution of selected characteristics of 319 colorectal cancer patients and 310 controls are shown in Table 2. Ages ranged from 20-85 years for cases and 18-89 years for controls (medians of 51 and 45 years, respectively Pvalue=0.000). The mean age of CRC patients and controls respectively were 51.92 (Standard deviation, SD±13.92) and 45.84 (SD±15.90) years. CRC patients showed significantly higher mean age in comparison with control groups (Pvalue<0.05). Logistic regression method was used to remove the effect of age, gender and smoking status variations as confusing factors. In addition to these, the distribution of clinicopathologic variables such as tumor location, grade, TNM and Dukes stage are shown in Table 2. Among the CRC cases, the rate of colon cancer was noticeably more than that of rectum (74.1% colon and 25.9% rectum location). According to histological differentiation of tumor grades, 9.5%, 25.2%, and 48.5%, patients were classified in three grades, poor, moderate, or well grade, respectively. Tumor grade distribution was such that well grade status was more common (48.5%) than others. Besides, patients were grouped into five classes from 0 to IV regarding tumor node metastasis (TNM) at the time of diagnosis, 1.3%, 13.1%, 38.0%, 38.0% and 9.5% respectively. Digestion of the PCR product of 257-bp with MseI enzyme results in 152 and 105-bp fragments for Lys allele and a single 257-bp fragment for Glu allele. Figure 1 displays the PCR-RFLP patterns of the K589E genotypes in EXO1 gene. The frequency distribution of the genotypes and alleles of the Exo1 K589E in CRC patients and controls are summarized in Table 3. The Glu/Glu, Glu/Lys and Lys/Lys genotypes were found in 145 (45.5%), 148(46.4%) and 26 (8.2%) of 319 colorectal cancer patients and in 147 (47.7%), 136 (43.9%) and 27(8.7%) of 310 controls. Neither genotypic distributions of patients nor controls deviated from those expected from Hardy-Weinberg equilibrium (Pvalue=0.164 and 0.572 respectively). These suggest that there was no population stratification and sampling bias. As revealed by these results, genotypic and allelic distribution of this genetic polymorphism of EXO1 was not significantly different between CRC patients and controls (OR=1.033, 95% CI=0.814 -1.312). Although there was no statistically significant association between alleles and disorder, Glu is the most allele in this population and its frequency was 68.7% in patients compared with 69.4% in controls.
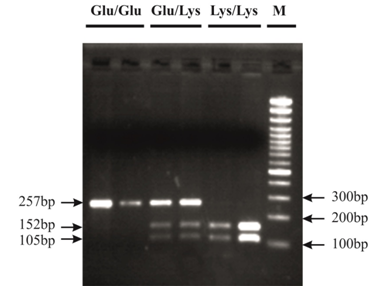
Figure 1 Analysis of EXO1 K589E polymorphism shown on 2.5% agarose electrophoresis. M: 100bp DNA size marker, Glu/Glu homozygous (257-bp), Glu/Lys heterozygous (257, 152, 105-bp) and Lys//lys homozygous (152 and 105-bp).
Characteristics |
Patients (n=319) |
Controls (n=310) |
pa |
||||
n |
% |
Mean (SD) |
n |
% |
Mean (SD) |
||
Age (Years) |
51.92±13.92 |
45.84±15.90 |
0 |
||||
Gender |
0.014 |
||||||
Male |
165 |
51.7 |
130 |
41.9 |
|||
Female |
154 |
48.3 |
180 |
58.1 |
|||
Smoking Status |
0.956 |
||||||
Never |
266 |
83.5 |
259 |
83.5 |
|||
Current |
53 |
16.6 |
51 |
16.5 |
|||
Tumor Grade |
|||||||
Well |
148 |
48.5 |
|||||
Moderate |
77 |
25.2 |
|||||
Poor |
29 |
9.5 |
|||||
Not Determined |
51 |
16.7 |
|||||
Location |
|||||||
Colon |
226 |
74.1 |
|||||
Rectum |
79 |
25.9 |
|||||
TNM Stage |
|||||||
0 |
4 |
1.3 |
|||||
I |
40 |
13.1 |
|||||
II |
116 |
38 |
|||||
III |
116 |
38 |
|||||
IV |
29 |
9.5 |
|||||
T |
|||||||
T0 |
4 |
1.3 |
|||||
T1 |
15 |
4.9 |
|||||
T2 |
38 |
12.5 |
|||||
T3 |
204 |
66.9 |
|||||
T4 |
24 |
7.9 |
|||||
Unknown |
20 |
6.6 |
|||||
N |
|||||||
N0 |
160 |
52.5 |
|||||
N1 |
92 |
30.2 |
|||||
N2 |
32 |
10.5 |
|||||
Unknown |
21 |
6.9 |
|||||
M |
|||||||
M0 |
276 |
90.5 |
|||||
M1 |
29 |
9.5 |
|||||
Dukes Stage |
|||||||
A |
16 |
15.2 |
|||||
B |
139 |
45.6 |
|||||
C |
122 |
40 |
|||||
D |
28 |
9.2 |
|||||
MSI |
|||||||
High |
27 |
25.2 |
|||||
Low |
15 |
14.1 |
|||||
Stable |
65 |
60.7 |
|
|
|
|
|
Table 2 Characteristics of colorectal cancer patient and control groups
ap> 0.05 for differences between patients and controls
Amino Acid |
Genotype |
Patients¶ (n=319) |
Controls¶(n=310 ) |
P* |
OR (95% CI) |
|
Crude |
Adjusted† |
|||||
K589E |
Glu/Glu |
145 (45.5 %) |
147 (47.7%) |
0.814 |
1 (Reference) |
1 (Reference) |
Glu/Lys |
148 (46.4%) |
136 (43.9%) |
0.556 |
1.103 (0.796-1.530) |
1.133 (0.810-1.584) |
|
Lys/Lys |
26 (8.2%) |
27 (8.7%) |
0.936 |
0.976 (0.544-1.753) |
0.994 (0.564-1.810) |
|
Glu/Lys and Lys/Lys |
174 (54.5%) |
163 (52.6%) |
0.621 |
1.082 (0.791-1.481) |
1.109 (0.804-1.530) |
|
Allele Frequencies |
||||||
Glu |
438 (68.7%) |
430 (69.4%) |
1 (Reference) |
|||
|
Lys |
200 (31.3%) |
190 (30.6%) |
0.788 |
1.033 (0.814-1.312) |
|
Table 3 Distribution of EXO1 genotypes among colorectal cancer patients and controls
¶The observed genotype distribution of patients and controls were in agreement with the Hardy-weinberg equilibrium.
*p values were for the difference in genotype frequencies between patients and controls.
†Ors were adjusted for age, gender and smoking status.
We analyzed the association between the EXO1 genotypes and clinicopathological characteristics in colorectal cancer patients. We found no evidence for association of rs1047840 with TNM and Dukes stage, tumor location and tumor grade (Table 4).
Characteristics |
Genotype |
|||
Glu/Glu |
Glu/Lys |
Lys/Lys |
PValue |
|
Tumor Grade |
0.523 |
|||
Well |
71 |
62 |
15 |
|
Moderate |
34 |
37 |
6 |
|
Poor |
15 |
14 |
0 |
|
Not Determined |
20 |
27 |
4 |
|
Location |
0.397 |
|||
Colon |
98 |
108 |
20 |
|
Rectum |
41 |
33 |
5 |
|
TNM Stage |
0.732 |
|||
0+I+II |
72 |
77 |
11 |
|
III+IV |
67 |
65 |
13 |
|
T |
||||
T0 |
3 |
0 |
1 |
0.291 |
T1 |
9 |
6 |
0 |
|
T2 |
18 |
19 |
1 |
|
T3 |
90 |
95 |
19 |
|
T4 |
8 |
13 |
3 |
|
Unknown |
11 |
9 |
0 |
|
N |
0.512 |
|||
N0 |
70 |
78 |
12 |
|
N1 |
46 |
36 |
10 |
|
N2 |
14 |
16 |
2 |
|
Unknown |
9 |
12 |
0 |
|
M |
0.254 |
|||
M0 |
130 |
125 |
21 |
|
M1 |
9 |
17 |
3 |
|
Dukes Stage |
0.521 |
|||
A |
9 |
6 |
1 |
|
B |
59 |
70 |
10 |
|
C |
62 |
50 |
10 |
|
D |
9 |
16 |
3 |
|
Table 4 Association between EXO1 genotypes and clinicopathological characteristics
TNM, tumor node metastasisTNM, tumor node metastasis
We also investigated the association between Exo1 genotypes and MSI status among tumors from 107 colorectal cancer patients. There were 27 (25.2%) MSI-H, 15 (14.1%) MSI-L and 65 (60.7%) MSS. Analysis of the data revealed that there were no significant correlation between Exo1 genotypes and MSI status (Pvalue= 0.536).
In order to find potential predisposing risk factors of CRC, we selected an important polymorphism of EXO1 gene and investigated whether the EXO1 K589E polymorphism could have an effect on susceptibility to CRC in an Iranian population. We also investigated the association of this polymorphism with clinicopathological features such as TNM stage, tumor grade and location.
We found, for the first time, that variant genotypes of K589E were not associated with risk of CRC. It is well-known that EXO1 is a unique exonuclease involved in human MMR system. It plays a critical role as both 5'-3' and 3'-5' nucleases, contributing to the overall integrity of the MMR complex.41,42 The MMR system plays a major role among the DNA repair systems, as it is responsible for correcting base-base mispairs and small insertion/deletion loops.13,41,43 Since this system plays a major role in maintaining the genome integrity, defection of DNA MMR function will lead to increase the frequency of spontaneous genetic mutations of oncogene, tumor suppressor and other genes, finally leading to tumor initiation and development. Regarding the important role of EXO1 in MMR system, this gene has become a famous target gene and widely investigated for its association with several cancers such as oral, CRC, lung and breast.17,35,37,44,47 In this regard, numerous studies have reported several SNPs of EXO1 as genetic risk factors of cancer.
In 2012, the association between T439M polymorphism of EXO1 and hepatocarcinogenesis was also examined in a Turkish population, indicating that this polymorphism does not have a major role in genetic susceptibility to hepatocarcinogenesis.47 Besides, our group also found a relation between L757P polymorphism and susceptibility to CRC in an Iranian population.12 In 2012, another study investigating Chinese population found that the A allele of K589E polymorphism increases the risk of cervical cancer.37 The EXO1 K589E (rs1047840) polymorphism is a non-synonymous SNP located on exon12 of the EXO1 gene. This polymorphism results in dramatic amino acid change at codon 589 from a negatively charged glutamate to a positively charged lysine residue, in turn, possibly affecting the protein functions. Also because Glu589Lys located at an exonic splicing enhancer (ESE) region, it might influence the production of EXO1 mRNA.48 Although some studies have suggested an association between K589E and several cancer types, our results demonstrated no significant relation between this polymorphism and susceptibility to colorectal cancer in Iranian population. Our results in line with Zienolddiny et al.,36 have reported no significant association of EXO1 K589E polymorphism and risk of non-small cell lung cancer in Caucasian Norwegian population.36 On the contrary in other reports SNPs of EXO1 K589E have been associated with risk of several other types of cancer. Jin et al.,48 found that K589E polymorphism is associated with the risk of lung cancer in a Chinese population. Similar conclusion was obtained in the study of Taiwanese population, when it was found that the A allele of this polymorphism has a significant association and increase risk of lung cancer (P=0.0097).35 Interestingly, in other Taiwanese studies, in 2009, Wang et al.,17 Tsai et al.,44 & Bau et al.,45 reported that the A allele EXO1 K589E conferred a significantly increased risk of gastric, oral and breast cancers respectively. In recent reports, SNPs of EXO1 K589E have been investigated with susceptibility to cervical cancer and HCC development. In 2012, Luo et al.,37 found a significantly different distribution in the frequency of genotypes between cervical cancer patients and controls. The A allele increased the risk of cervical cancer (OR: 1.67; 95% confidence interval, 1.13-2.45) in Chinese patients.37 At that time, Bayram et al.,38 found that Lys/Lys genotype of the EXO1 K589E polymorphism is associated with increased risk of HCC development in Turkish population. Therefore, it can be speculated that this difference in distribution frequency of EXO1 K589E genotypes may be due to the differences in the pathways of carcinogenesis among different tissues. Moreover, these discrepancies may be attributable to the differences of the ethnic variation. For instance, according to International HapMap database, Glu allele frequency of EXO1 K589E polymorphism among different ethnicities is as follows:
0.57 in Africans, 0.62 in Northern and Western Europeans, 0.80 in Chinese and 0.76 in Japanese (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36).38 Our results confirmed that the Glu allele frequency is the most common one in Iranian population like other countries. Also our results didn't show any significant correlation between clinicopathological features, MSI status and EXO1 genotypes. According to our recent research, some previous studies investigated the influence of SNPs and clinicopathological features of CRC. For instance, Mates et al.,49 found an association between rs293827 and rs3802842 of SMAD7 gene and site specific difference of CRC. Their results revealed that increased susceptibility to development of rectal cancer rather than colon has been shown among carriers of risk allele at these loci. C allele carriers at rs3802842 are associated with a lower risk of rectal tumors. Our results are in concordance with few studies. For example Yamamoto et al.,21 didn't find any significant association between EXO1 T439M, P757L polymorphism and clinicopathological characteristics such as MSI status, tumor location, grade and TNM stage. Also Yoshiya et al.,50 didn't discover a significant difference between TNM stage, tumor location, tumor grade and EXO1 P757L genotypes.
Some strengths and limitations of this study should be addressed. The most important strength is its well-defined homogenous sample with detailed clinical data. In addition, in the case and control groups, the agreement between the observed distributions of EXO1 K589E genotype frequency with the expected according to the Hardy-Weinberg equilibrium suggested no selection bias. The limitations of our study are as follows. First limitation of the present study is sample size; considering the moderate sample size (319 cases and 310 controls) in this study, the genotype differences may be attributable strictly to chance. Second, this study only focused on one polymorphism of the EXO1 gene without taking in to consideration gene-gene interactions and interactions between different loci on the same gene, which may affect the risk of CRC. Finally, the potential deviation of information may be due to hospital-based case-control study design. Therefore, the possibility of chance finding could not be completely ruled out. Nevertheless, we should not exclude the possibility of true results.
The present study indicated, for the first time, that the EXO1 K589E polymorphism does not have any major role in genetic susceptibility to CRC at least in the studied population. Moreover this polymorphism may not be a suitable genetic predisposing biomarker for prediction of CRC clinicopathological features. However, further independent studies with sufficiently larger population are required to validate this result and clarify the unsolved issues. Understanding the biological mechanism of this polymorphism can be appeared by functional analysis and can lead to targeted prognosis and development of novel therapeutic strategies.
This study was supported by Gastroenterology and Liver Diseases Research Center, Shahid Beheshti University of Medical Sciences. The authors are very grateful to all participants for taking part in this study, all fieldworkers, data and laboratory staffs.
Authors declare that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.