eISSN: 2577-8307


Research Article Volume 3 Issue 2
Federal College of Forest Resources Management, Forestry Research Institute of Nigeria, Nigeria
Correspondence: Aghimien Ehimwenma Victor, Federal College of Forest Resources Management, Benin City, Edo State, Forestry Research Institute of Nigeria (FRIN), Nigeria
Received: March 08, 2019 | Published: March 19, 2019
Citation: Aghimien EV. Above-ground carbon stock estimation using pleiades satellite imagery of the secondary forest ecosystem in Ibadan, Nigeria. Forest Res Eng Int J. 2019;3(2):46-54. DOI: 10.15406/freij.2019.03.00077
Secondary forest ecosystem contributes to global climate change mitigation through carbon sequestration. Above-Ground Biomass (AGB) is the major component for monitoring and estimating Carbon Stocks (CS) and fluxes in tropical forests. However, information on Above-Ground Carbon Stock (AGCS) for the International Institute of Tropical Agriculture (IITA), which hosts relics of the undisturbed secondary forest ecosystem in south-western Nigeria, has not been documented. Therefore, AGCS of the secondary forest ecosystem was estimated using remote sensing techniques. Pleiades satellite data were used for this study. One hundred and forty plots of 50m x 50m were laid in IITA secondary forest using systematic sampling technique at 10% sampling intensity. Pleiades satellite imagery was acquired using Remote Sensing (RS) technique and spectral data for each sample plot extracted. The spectral indices used for AGB estimation were: Normalised Difference Vegetation Index (NDVI), Difference Vegetation Index (DVI), Infrared Percentage Vegetation Index (IPVI), Optimised Soil Adjusted Vegetation Index (OSAVI) and Re-normalised Difference Vegetation Index (RDVI). Regression equation was used for the prediction of AGB from where the total CS estimate was obtained. Data were analysed using descriptive statistics and linear regression analysis.
The AGB and CS ranged from 101.06 to 881,834.92 kg/ha and 50.53 to 440,917.46 kg/ha, respectively. The DVI had the highest AGB value which ranged from 187 to 15,577 kg/ha, followed by IPVI, RDVI and OSAVI which ranged from 7,561 to 12,324 kg/ha, 64.0591 to 133.178 kg/ha, 0.0134 to 0.5621 kg/ha, respectively, while NDVI had the least values which ranged from -0.01 to 0.48 kg/ha. The best AGB estimation model was AGB = exp. (3,496.61 + 0.99 x (RDVI) 1/2); Coefficient of Determination (R2) = 0.93, Bayesian Information Criterion (BIC) = 82.34). The total carbon stock ranged from 11,035 to 18,774 kg/ha. Model with re-normalized difference vegetation index was most suitable among other indices for estimating above-ground carbon stock. Therefore, effective integration of different sensor data will be an important research topic for improving above-ground biomass estimation performance.
Keywords: carbon stock prediction, secondary forest biomass, vegetation spectral indices, remote sensing
Remote Sensing (RS) measures have been applied to establish for natural forest resources management.1 Currently, remote sensing is extensively being used to collect data on forest Above Ground Biomass (AGB) and vegetation structure as well as to monitor and map vegetation biomass and productivity on large scales by determining the spectral reflectance of the vegetation structure.2 Remote sensing data are now considered to be the most dependable techniques of estimating spatial biomass in tropical forest over large areas. The technology has been applied to AGB estimation over large areas at a reasonable cot and with acceptable precision based on repetitive information collection with negligible effort.3 Several information sources can be used to map and estimate AGB for different land-cover categories; one of such method is the use of satellite images to estimate the attributes of forests through direct correlations or stepwise regression analyses with Vegetative indices, spectral bands or band ratios.4 Vegetation indices have been used to estimate AGB in several studies.5 A combination of information sources can be used to forecast vegetation structure variables over large expanse of lands.6 A hybrid classification approach combined with geographical information systems (GIS) analysis has been employed to expand land use mapping for Landsat data.7
There are uncertainties with regard to the precise estimation of above-ground biomass in the secondary forest ecosystems. In addition, the choice of techniques that can standardize and improve the accuracy of such estimates is far from being decisive. These uncertainties are accountable for overestimation or underestimation of above-ground biomass, typically ascribed to the complexity of the secondary forest ecosystems.8 These increased the problems of deriving secondary forest parameters mainly in those areas located in tropical and subtropical regions.9 It is generally recognized that information acquired with remote sensing techniques can play a significant role in estimating greenhouse gases (GHG) emission from secondary forest ecosystem changes. However, there is no known documented research that focused on a large area using remote sensing to estimates greenhouse gases.8 Conservatively, above-ground biomass estimation in forests involves cutting, which contributes to the reduction of the carbon sink size. Hence, assessing carbon from RS information may contribute positively to the mitigation of global warming. Thus, the essence of this study is to employ pleiades satellite imagery in estimating carbon sequestered in above-ground carbon stock of the secondary forest ecosystem in Ibadan, Nigeria.
The study area
Location
The International Institute of Tropical Agriculture (IITA) Secondary Forest Ecosystem lies at 07o 30' 0''N and 03o 53' 30''E and approximately 227m altitude in the city of Ibadan. Ibadan lies in the rainforest zone to the south and savanna zone to the north. To the west lies the Dahomey Gap, where savanna reaches almost to the coast in neighbouring Benin Republic. This arid zone form a barrier that several rainforest species are unable to cross, thus dividing West African Rainforest into two types, with upper Guinea forest to the West and Guinea-Congo forest to the East.10
Vegetation
The secondary forest ecosystem is known for its trees, climbers, shrubs, lianas and grasses. The forest area is classified as dry semi-deciduous rainforest, with a mixture of fast growing pioneer tree species, such as Anthocleista vogelii , Albizia spp., Ceiba pentandra, Newbouldia laevis, and interspersed with slow growing emergents, including Antiaris africana, Milicia excelsa, and Triplochiton scleroxylon, together with abundant climbers and lianas, especially of the genera Combretum and Dioscorea, and an understorey of shrubs such as Mallotus oppositifolius, Chassalia kolly and Sphenocentrum jollyanum (Figure 1).10
Laying of sample plots
Systematic sampling technique was used to select temporary sample plots (TSPs) of 50m x 50m (0.25ha) in size. The sample plots were primarily established with 10% sampling intensity. The total area of IITA secondary forest ecosystem was 350ha. One hundred and forty (140) sampling plots were systematically laid in the secondary forest ecosystem. Plots need to be allocated systematically so as to achieve above-ground biomass.11 The number of sample plots and the distance between plots were determined by the formula;12
N= TA X SiPs X 100 1
Where N = number of sample plots, PS = Plot size, TA = Total area of the forest and Si = Sampling intensity, while the distance between plots was determined by the formula:12
D= √(Af X 10000N) 2
Where D = inter plots distance (m), N = number of plots and Af = Area of the forest (ha).
50m
Estimation of AGCS within a sample plot
The mean plot AGB for each tree species in the secondary forest ecosystem of International Institute of Tropical Agriculture was computed and then multiplied by 0.25 (the number of 50m x 50m plots in a hectare) to acquire the AGB per hectare. However half of the value gave the AGCS per hectare for the secondary forest ecosystem.10
Satellite data collection on land-use and land-cover
Satellite data and pre-processing
Satellite data (Landsat 7 ETM) on past and historical land use and land cover patterns were obtained from GIS and database unit at the International Institute of Tropical Agriculture. Primary data was acquired from the most recent Pleiades sensor with a resolution of 50cm which was launched on the 2nd December, 2012 by AIRBUS Defence and Space in France.
Ground truthing
Field observations were carried out in the study area in order to authenticate information acquired from GIS and information base unit, IITA and the satellite imagery showing the secondary forest-cover.
Extraction of the study sites
The pleiades imagery was designed to collect unique information by combining very high spatial resolution with observation spectral bands in the visible and near-infrared as well as the short wave infrared. This new sensing platform is similar to other high resolution satellite such as WorldView-3 with two important additions. The pleiades imagery platform is well positioned to provide high resolution data, timely, and accurate insight to multiple applications, such as agricultural mapping, mineral exploration, and urban forestry monitoring.
Forest-cover classification technique
The Universal Transverse Mercator projection imagery was geo-rectified using image registration. Pleiades imagery bands were stacked, and the image was subset for the secondary forest ecosystem. The forest cover map was classified into non-forest, less dense and highly dense vegetation to estimate the AGB for each class using ERDAS imagine software. The classifications with trees species was analyzed using a hybrid classification technique with ArcGIS 9.1.13 The training patterns were developed through hybrid classification with the use of an unsupervised classification followed by a supervised classification. For the unsupervised classification, a K-means clustering algorithm was used to search for natural groups of pixels called clusters, which was located in the information by assessing the relative locations of the pixels in the feature space for separations between non-forest, less dense and highly dense vegetation features. The highly dense vegetation features were also identified for forest inventory verification in the study area. The maximum likelihood technique for the supervised classification was applied using analyst-defined training areas to determine the features of the forest cover type in the study location.
Vegetation indices (VIs)
Research have shown how the combinations of the measured reflectance properties at two or more wavelengths reveal specific non-forest, less dense and highly dense vegetation features, also known as Vegetation Indices.14–17
IPVI=NIR/NIR+red 3
DVI=NIR−red 4
(NIR–red)/(NIR + red) 5
OSAVI=((NIR–red)/(NIR + red +L))*(1 + L) 6
Where; L is a correction factor that equals 0.16 (the same equation as SAVI but with a correction factor of 0.16 instead of 0.5).18
RDVI=(NIR–red)/(NIR+ red)1/2 7
There are more than 150 existing Vegetation indices, with additional indices emerging as sensors advance and provide new information.19
Evaluation of Selected Models
The evaluation of the model was based the goodness of fit statistics such as Coefficients of determination (R2), Root mean square error (RMSE) and Bayesian information criterion (BIC).
Coefficient of determination (R2): It provides a measure of how observed outcomes are replicated by the model, as the proportion of total variation of outcomes explained by the model.20
R2=1− (∑ni=1(yi-i)2/n∑ni=i(yi−i)2) 8Where;
ˆyi = observed value, yi = predicted value, n = number of observation
Root mean square error (RMSE): It represents the sample standard deviation of the differences between predicted and observed values. It estimates must be low as much as possible in order to reduce biased.20
RMSE=√1n∑ni=1(yi− ˆyi)2 9
The significant differences between estimated and actual values were calculated to identify the best estimator.
Sx1x2 = √12(S2x1+S2x2) 10
Where:
ˉX Sx1x2 =Means for predicted and observed data respectively
= Pooled standard deviation
Bias is a measure of the mean difference between observed and predicted values. The smaller the differences the better the model fits the calibrated data.
Bias= ∑ni=1(yi−ˆyi)/n 11
Where;
ˆyi = observed value, yi =predicted value, n = number of observation
Bayesian information criterion (BIC): It is a criterion for equation selection among a finite set of equations; the model with the lowest BIC is preferred for model development. It is based, in part, on the likelihood function and it is closely related to the Akaike information criterion (AIC). The mathematical expression for comparing maximum likelihood equations is defined as;
BIC=−2*In (∑ni=1(yi− ˆyi)2/n)+In(N)*K 12
Where;
yi = observed value, ŷi = predicted value, N = number of observations and K = number of parameters estimated.
Root Mean Square Error (RMSE) value fitting of criteria to estimate evaluation is most important and forecasts are increasingly accurate with lower RMSE values.
Cross-validation is a method that removes each measured location one at a time in order to predict their values on the basis of the measured values in the entire dataset. It is essential to validate the models selected after assessment before their suitability can be introduced to forest resource management. Validation was carried out by comparing the models’ output with values observed on the field. Field data was divided into two sets. The plot data was randomly divided into calibration (70% of the data) and validation (30% of the data) data sets with reference to past Ikonos satellite imagery. The validation was tested by using scatter plots for residuals between observed and predicted values. Cross-validation criteria provide values such as Standard Error (Mean), which assess the accuracy of the chosen model. The Standard Error (Mean) values were used to assess the variability in the predictions from the measurement values. The Standard Error (Mean) values should be low, this identify that predictions are close to its true values.
Standard Error (Mean): It is the mean standardized error that can be calculated by the following equation:
Standard Error (Mean)=1n[n∑i=1(yi−ˆyi)σi] 13
Where Yi = the observed values, ˆyi = the predicted values σi = Standard error and n = number of observations.
Model selection criteria
Above-Ground Carbon Stock Estimation
The descriptive statistics of the examined forest inventory and remote sensing variables for the 140 sampling plots are presented in Table 1. There were variation in mean remote sensing variables across IITA secondary forest ecosystem, which ranged from 0.35 to 16903.62 with lower values for standard deviation which ranged from 0.02 to 1428.61, and the variance had values that ranged from 0.00 to 2040912,which means that tree species were well distributed as some sample plots still require regeneration. The coefficient of variation value is the most discriminating factor for describing variability; when coefficient of variation is less than 10 %, it shows small variability but when coefficient of variation is more than 90 %, it shows high variability. In these study, the coefficient of variation ranged from 5% to 35%, indicating variability in IITA secondary forest ecosystem as revealed in Table 1. The Normalized Difference Vegetation Index (NDVI) digital map values for this study ranged from -0.0115883 to 0.484797, with the higher value representing ‘greener’ surfaces while values less than zero typically do not have any ecological meaning, so the range of the index is truncated to 0.0 to +1.0.21 Very low value of NDVI corresponds to barren areas of rock, sand etc. Moderate values represent shrub and grassland as presented in Figure 2A. Many factors could influence NDVI values like plant photosynthetic activity, biomass, total plant cover, plant and soil moisture, plant stress as well as time of day or season when the images were acquired.21 The Difference Vegetation Index (DVI) digital map values ranged from 187 – 15577, with the higher value reflecting very healthy vegetation in the near infrared part of the spectrum, which is invisible to human eyes. High DVI value of 15577 corresponds to dense vegetation such as that found in IITA secondary forest at their peak growth stage as shown in Figure 2B. As plant canopy changes from early raining-season growth to dry-season maturity and senescence, these reflectance properties also change properties.21
Stats |
AGB |
NDVI |
DVI |
OSAVI |
IPVI |
RDVI |
Min. |
21.16 |
0.18 |
4315 |
0.21 |
1 |
91.18 |
Mean |
105.73 |
0.35 |
8758.47 |
0.40 |
1.73 |
106.75 |
Max. |
211.58 |
0.44 |
12285 |
0.51 |
2 |
120.94 |
SD |
37.33 |
0.05 |
1428.61 |
0.06 |
0.45 |
5.23 |
Variance |
1393.80 |
0.00 |
2040912 |
0.00 |
0.20 |
27.35 |
Se (mean) |
3.16 |
0.00 |
120.74 |
0.00 |
0.04 |
0.44 |
CV |
0.35 |
0.14 |
0.16 |
0.14 |
0.26 |
0.05 |
Table 1 Descriptive Statistics for Spectral Variables
Key: AGB, above-ground biomass; NDVI, normalised difference vegetation index; DVI, difference vegetation index; OSAVI, optimised soil adjusted vegetation index; IPVI, infrared percentage vegetation index;
RDVI , renormalized difference vegetation index; Max, maximum; min, minimum; SD, standard deviation; Se ,standard error; CV, coefficient of variation
The Infrared Percentage Vegetation Index (IPVI) digital image shows a high value of 12324, meaning IITA secondary forest ecosystem has healthy vegetation while for the low value of 7561 had less healthy vegetation with white and brown patches, which represented sparse vegetation as shown in Figure 3A. The more leaves a plant has, the more these wavelengths of light are affected. The Optimized soil adjusted vegetation index (OSAVI) digital image analysis with a value of 0.562119 shows a high vegetation cover for the canopy background while 0.0134307 comprised of area with low vegetation cover of soil variation as presented in Figure 3B.
The Renormalized difference vegetation index (RDVI) value of 133.178 obtained represents healthy vegetation while 64.0591 recorded indicates area with low healthy vegetation cover as revealed in Figure 3C. This is an indication that if concerted efforts to plant more trees are not quickly made to vegetation coverage of IITA secondary forest ecosystem; it will no longer be supportive in providing services such as protection of the environment.
In Table 2, the parameters of the models fitted to above-ground biomass data are presented (Equations 14 - 16). Then, the best-fitting equation for each variable as judged by the R2 value and significance were used for the transformations. The transformed variables were then used to perform the subsequent model. Finally, fitting the results of the exponential function was used to model the above-ground biomass. The result shows that the model has an R2 value of 0.93 for RDVI. The model forms are as follows;
Exponential function: y= b0eb1x 14
Logarithmic function: y= b0+ b1 lnx 15
Power function: y= b0xb1 16
Where;
y = dependent variables
bo and b1 = coefficients
x = independent variable
ln = log normal
e = error
Models |
Parameters |
R2 |
BIC |
p-value |
RMSE |
|
|
b0 |
b1 |
|
|
|
|
Power function |
254.20 |
1.22 |
0.91 |
87.55 |
0.0000 |
32.66 |
Logarithmic function |
2.34 |
-143.98 |
0.90 |
99.02 |
0.0001 |
33.40 |
Exponential function |
3496.61 |
0.99 |
0.93 |
82.34 |
0.0000 |
31.39 |
Table 2 Best estimated model parameters and performance criteria
The starting value of each parameter was iteratively set and the optimum value with the smallest sum of squared deviations was chosen as the final parameter estimates. Convergence was achieved in all the fitted models. Each equation was evaluated using the coefficient of determination (R2), Bayesian information criteria (BIC), and Root mean square error (RMSE). The results of the evaluations are presented in Table 2, which also shows estimated model parameters. The R2 of the models ranged from 0.90 to 0.93, BIC ranged from 82.34 to 99.02 while RMSE ranged from 31.39 to 33.40. Exponential function produced the best fit with the lowest R2, BIC and RMSE, but with only small differences from models 1and 2 had the poorest fit as presented in Table 2. Above-ground biomass (AGB) may be directly estimated using linear or nonlinear regression equations, which can be better derived from remotely sensed information.21 A combination of spectral responses and image textures has proven useful in improving AGB estimation performance. The incorporation of remote sensing and GIS data was also useful in improving AGB estimation results when multi-source information are obtainable. In practice, the lack of sufficient and high-quality AGB sample plots is a major difficulty in developing AGB estimation models and in implementing precision and validation. The exponential function relating spectral variables to AGB was superior to the other functions. Each equation was evaluated using the root mean square error (RMSE), coefficient of determination (R2) and probability value. Equations involving pleiades predictors were developed, and fitted to the calibration data set, using the nonlinear least square technique which simultaneously eliminated variables which were not significantly contributing to the prediction of AGB.
All the spectral variable models obtained in this study are presented in Table 3, using vegetation indices as the independent variables.
The best AGB for each of the spectral variables are:
AGB= exp.: (3496.61+0.99 x (NIR–red)/(NIR+red)1/2) Model 1
S/N |
Regression Model |
R2 |
p-value |
RMSE |
Bias |
1. |
AGB = exp.: (3496.61 + 0.99 x (NIR – red)/(NIR + red)1/2) |
0.93 |
0.0000 |
31.39 |
-0.0042 |
2. |
AGB = exp.: (3496.61 + 0.99 x (NIR – red)/(NIR + red)1/2) |
0.93 |
0.0000 |
31.39 |
-0.0042 |
3. |
AGB = exp.: (45.21 + 1.00 x (NIR – red)) |
0.92 |
0.0000 |
34.48 |
2.1344 |
4. |
AGB = exp.: (25.94 + 30.88 x ((NIR – red)/(NIR + red + 0.16))x(1 + 0.16) |
0.92 |
0.0000 |
32.71 |
-0.0538 |
5. |
AGB = exp.: (57.54 + 1.41 x NIR / NIR + red |
0.85 |
0.0000 |
40.35 |
2.9639 |
Table 3 Non-Linear Equations for the Estimation of AGB
Key: Model 1, RDVI; Model 2, NDVI; Model 3, DVI; Model 4, OSAVI; Model 5, IPVI
The selected model was significant at α = 0.05 probability level and a coefficient of determination (R2) value of 0.93, the first model (Model 1) had moderate root mean square error of 31.39 with a bias of -0.0042. Model 1 was selected as the best model for predicting AGB in IITA secondary forest ecosystem. When NDVI was used as the explanatory variable, model 2 contributed a high R2 value of 0.92 with a moderate root mean square error of 32.71 and a bias value of 0.3527. The model is stated below;
AGB= exp.: (25.92 + 53.40 x (NIR–red)/(NIR+red)) Model 2
It implies that model 1 did not violate the assumptions of a non-linear regression equation, which indicates that the distribution of variance of the explanatory variable should be constant for all values of the explanatory variable as it makes the data cluster perfectly along the regression line. The introduction of DVI as an explanatory variable for predicting AGB, model 3 contributed a high R2 value of 0.92 with a moderate root mean square error of 34.48 and a bias value of 2.1344. The expression of the model is represented as;
AGB = exp.: (45.21+1.00 x (NIR – red)) Model 3
With the introduction of OSAVI as another explanatory variable for predicting AGB, model 4 contributed a high R2 value of 0.92 with a moderate root mean square error of 32.71 and a bias value of -0.0538. The model is expressed as follows:
AGB = exp.: (25.94 + 30.88 x ((NIR – red) / (NIR + red + 0.16))x(1 + 0.16) Model 4
A modest improvement was found in the root mean square error by adding OSAVI as a predictor variable, which accounts for differences between AGB and vegetation indices of similar characteristics. A major factor that affects the accuracy of above-ground biomass estimation in one study area may become minor in another. Therefore, spatial uncertainty analysis and error budget should be conducted to identify the major factors and further to provide a mechanism of quality control for forest above-ground biomass equation predictions.22 Many authors have also investigated the uncertainties in estimating and mapping forest above-ground biomass. For example, Chave,23 reported that uncertainty in forest above-ground carbon estimations was greater than the changes in carbon sequestration via forest management and planning. Saatchi et al.,24 found uncertainty of about 20% when regression equations are used for total forest above-ground biomass mapping.
Adding IPVI as an independent variable for predicting AGB, model 5 revealed a high R2 value of 0.85 with a moderate root mean square error of 40.35 and a bias value of 2.9639. The model is expressed as;
AGB = exp.: (57.54 + 1.41 x NIR / NIR + red Model 5
In some fast growing tropical tree species, forest cover has been reported to increase with age. Model 5 as a predictor for predicting AGB was therefore not adequate for predicting above-ground bole biomass.
Estimation of total carbon stock map
The developed best equation was further applied to ArcGIS to produce an estimate of the total carbon stock map that ranged from 7,788 –11,035kg/ha and 11,035 – 18,774 kg/ha, indicating that the greener area represented healthy vegetation, the moderately green area signifies shrubs. The high carbon stock value of 18,774 kg/ha, indicate the presence of moderate carbon content in the study area as presented in Figure 4. Chen et al.,19 reported that the precise estimation and mapping forest above-ground biomass is critical to formulating global and national strategies to mitigate carbon concentration in the atmosphere and consequently global change in climate.
Predict future land cover vegetation map
All the land cover categories are affected by climate change which could be in the positive or negative direction. Positive value of net change refers to decrease in size of the land cover category while negative change corresponds to expansion in size of the land cover classification. The spatial distribution of land vegetation map in IITA secondary forest ecosystem displays a heterogeneous distribution of AGB of non-forest, less dense vegetation and highly dense vegetation, respectively. The acquired land cover imageries namely; Landsat 7 ETM of 1986 and Pleiades of 2016 in the secondary forest ecosystem were analysed using markov chain to produce an image differencing as presented in Figure 5. The land cover vegetation map was further projected for 2047 in the secondary forest ecosystem as shown in Figure 6. Assuming the total land areas of IITA secondary forest ecosystem is covered by trees, it would mean that large amount of above-ground carbon would be stored probably running into billion tons and this would have great implication on above-ground biomass that would be sequestered from the environment. This would in turn mitigate the effect of CO2 being emitted from other sources especially fossil fuel burning and farming. The probability that non-forest will remain is 50% in the next 30 years while the probability that the non-forest will change to less dense vegetation is 5% and the probability that the non-forest will change to highly dense vegetation is 2%. None-forest coverage are potential areas for enhancement through assisted regeneration and enrichment planting. The probability that the less dense vegetation will change to non-forest is 50%, while the probability that the less dense vegetation will not change is 42%. This may imply that the growth of small trees and loss of bigger trees over the time period. The probability that the less dense vegetation will change to highly dense vegetation is 8%. The probability that the highly dense vegetation will change to non-forest is 0%, while the probability that the highly dense vegetation will change to less dense vegetation is 52% and the probability that the highly dense vegetation will not be disturbed is 89% as shown in Table 4.
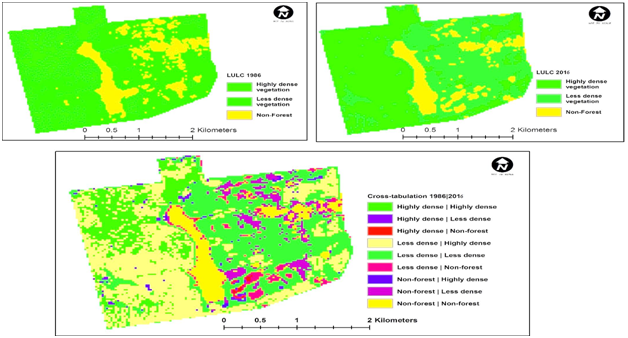
Figure 5 Transition probability map derived from land–cover maps of 1986 and 2016 for IITA forest reserve.
|
Class 1 |
Class 2 |
Class 3 |
Class 1 |
0.4961 |
0.5039 |
0.0000 |
Class 2 |
0.0534 |
0.4224 |
0.5242 |
Class 3 |
0.0209 |
0.0845 |
0.8947 |
Table 4 Transition Probability Matrix of Land-Use and Land-Cover Change
Key: Class 1, non-forest; Class 2, less dense vegetation; Class 3, highly dense vegetation
Evaluation and validation of the selected model
For the purpose of testing the developed equations, quantitative tests were used for the model evaluation. In each model, the observed values were assessed against the predicted values. The significant variance ratio (F) at 5% probability level, best line of fit with high coefficient of determination (R2), model function and Bayesian Information Criterion (BIC) were used as a guide for assessing the model suitability while the root mean square error (RMSE), residual plots and normal probability plots were used as an indicators of the model predictive ability. When all the model functions were tested for RDVI as explanatory variable for predicting AGB, the exponential model had the best BIC value of 82.34, followed by logarithmic function with a value of 99.02 and power function with a value of 87.55 as presented in Table 2. The evaluation of the best exponential model was considered using the highest value of coefficient of determination (R2), low values of RMSE, residual plots and normal probability distribution of plots (Table 2). Five models were developed using exponential function, model 1 was selected as the best for estimating AGB with a high R2 value of 0.93 and moderate RMSE value of 31.39 (Table 3).
Cross-validation is a method that removes each measured location one at a time in order to predict their values on the basis of the measured values in the entire dataset. Further validation of model 1 was carried out to examine the performance of the models in estimating AGB as an independent data set, outside the data used for building the models with past reference map (Ikonos imagery). The plot data was randomly divided into calibration (70% of the data) and validation (30% of the data) data sets. The validation was tested by using scatter plots for residuals between observed and predicted values. The model was applied to the validation data set and the predicted AGB of the validation plots compared to the observed AGB of the plots. It was observed that the predictions by the model agree closely with the actual observations. The validation result of standard error (Mean) had a value of 2.45 while the coefficient of variation had a value of 0.13 (Table 5). Therefore model 1 is regarded suitable for practical estimation of AGB and the validation map of IITA secondary forest ecosystem (Figure 7).
Variable |
Min |
Mean |
Max |
SD |
Variance |
CV |
Se (mean) |
Skewness |
kurtosis |
AGB |
93.21 |
120.18 |
174.02 |
15.88 |
252.29 |
0.13 |
2.45 |
0.96 |
4.75 |
Table 5 Descriptive statistics of data validation
Where: SD, Standard deviation; Min., Minimum; Max, Maximum; CV, Coefficient of variation; Se (mean), Standard error (mean).
In summary, probability that the highly dense vegetation of IITA secondary forest ecosystem not changing would be factual because for this to occur, it means the study area would be free of disruptive human activities for several years possibly running into years, if not decades in order for these study area to revert to its original state which may be practically impossible considering the agricultural activities. However, the management of IITA had not been consistent with enrichment planting. Model with re-normalized difference vegetation index was most suitable among other indices for estimating above-ground carbon stock. Therefore, effective integration of different sensor data will be an important research topic for improving above-ground biomass estimation performance. However, it should be noted that remote sensing techniques does not measure above-ground biomass directly, but rather it measures spectral reflectance of trees. Therefore, precise above-ground biomass estimation is necessary for improved understanding of deforestation impact on environmental degradation and global warming.
None
The author declares there are no conflicts of interest.

©2019 Aghimien. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
This is a modal window.