In many health studies, powerful tools for the statistical analysis are the survival analysis techniques that could be useful, for example, to identify risk factors or treatments that influences the survival or cure probability of a certain disease. In general, survival analysis consists in a set of techniques and statistical models commonly used when the random variable of interest is the time until the occurrence of a specific event, such as the time until the occurrence of a disease or the time until the patient’s death. A concept that differs survival analysis from others statistical analysis is the presence of censored data that occur when we have partial individual information about the time of occurrence of the variable of interest, however we do not know the exact time of occurrence of the event, that is, the real time of occurrence may exceed the observed time. The censored data can occur for a variety of reasons as the loss of monitoring of the patient over time and the non–occurrence of the event of interest until the end of the experiment. According to Colosimo and Giolo1 there are two reasons that justify the use of censored data in statistical analysis: (I) although these observations are not complete, they provide information about the patient’s lifetime and (II) the omission of the observations censored can lead to the calculation of biased estimates.
In survival analysis, usual parametric and non–parametric tools are widely used for analyzing data from time to event data. These tools are useful when some observations are censored and the event of interest was not seen in all patients during the follow–up period. The most used procedures include the mortality table, the Kaplan–Meier estimator for the survival function, the Cox proportional hazards model and parametric survival models. Parametric models are more flexible than Cox proportional hazards model, especially when there is no proportionality of risks between groups and are based mainly on two important functions, the survival function and the hazard function. These techniques are described in several textbooks as Kalbeisch and Prentice,2 Klein and Moeschberger3 and Kleinbaum and Klein.4
Let a non–negative random variable T related to the failure time, then the survival function is defined as the probability that an observation will not fail until a certain time t, that is, the probability that an observation will survive time t. In probabilistic terms,
On other hand, the hazard function
represents the instantaneous failure rate at time
conditional on survival time
and is very useful to describe the distribution of patient’s lifetime. In probabilistic terms,
Nonetheless, a common situation in many lifetime studies, particularly in cancer research, occurs when it is expected that a fraction of individuals will not experience the event of interest, this fraction of individuals often are immune or are cured. The presence of immune or cured individuals in a data set is usually suggested by a Kaplan–Meier plot of the survival function, which shows a long and stable plateau, with several censored date at the extreme right of the plot.5–8 However, an efficient and commonly applied technique is to consider a mixture of two populations, one susceptible to the event of interest adopting a base probability distribution to model the survival time of susceptible patients, and one of the most common distributions as, for example, the Weibull distribution9–11 for the non–susceptible population.
According to Hjorth,12 one or two parameters distributions have some important limitations such as the inability to model data that presents a bathtub risk function, for example. However, the most flexible distributions and with the largest number of parameters, may have inaccurate estimates, when there is a small sample size. In this way, this paper present a mixture cure rate model based on the Mirra distribution13 to estimate survival and hazard curves. The choice of Mirra distribution is justified due it number of parameters (two–parameters) and the bathtub shapes for the hazard function. Two dataset sets are considered to illustrate the proposed methodology. The first, related to gastric cancer, presents the bathtub shape for the empirical hazard function; and the second, related to breast cancer, presents a decreasing shape for the empirical hazard function.
The paper is organized as follows: in Section 2, it is presented the mixture Mirra cure rate model as its likelihood function. A brief description of the Bayesian approach and some discrimination criteria are also presented in Section 2. Section 3 presents the statistical analysis based on mixture Mirra cure rate model for two cancer data: gastric and breast cancer. The estimation of the survival and hazard curves are also presented in Section 3. Finally, Section 4 close the paper with some concluding remarks.
The mirra distribution
A first approach of the Mirra distribution (TPM) was introduced in the literature by Subhradev Sen.13 These authors studied most of it properties and illustrate how the Mirra distribution was synthesized as a special finite mixture of exponential and gamma distributions. In this way, let
be a continuous random variable following Mirra distribution with parameters
and
. The probability density function (pdf) and survival function (sf) of the random variable
is given, respectively, by,
(3)
and,
(4)
where
and
. Thus, the hazard function (hf) is represented by,
(5)
As noted by Subhradev Sen13 the hf of this distribution has the shape of a bathtub, decreasing to
and increasing to
. Moreover, these authors also showed that the hf is limited with the following limits,
(6)
In Figure 1, it is illustrated the shape of pdf, sf and hf. From those plots, it is possible to see the bathtub shape for the hf (green line). Also, for example, assuming
(green line), we have that the limits of the hf are given by
.

Figure 1 Behavior of the pdf (left panel), sf (middle panel) and hf (right-panel) for TPM distribution assuming arbitrary values for the parameters
and
.
A great advantage for the use of TPM distribution is that the special cases maintain the bathtub shape for the hf. These cases are described by,
- When we consider
, we get the Mirra–silence distribution (MSI), with probability density function and survival function respectively given by
(7)
- In case of
, is obtained the Mirra–surrender distribution (MSII), with probability density function and survival function respectively given by
(8)
- If we consider
, we have in this case the the xgamma distribution (XG), proposed by Sen et al.,14 with probability density function and survival function respectively given by
(9)
Mixture mirra cure rate model
Let us denote by
the event of interest. Following Maller and Zhou,15 the standard cure rate model (or mixture cure rate model) assuming that the probability of the time–to–event to be greater than a specified time
is given by the survival function,
(10)
where
is the mixing parameter which represents the proportion of “long–term survivors”, “non–susceptible” or “cured patients”, and
denotes a proper survival function for the non–cured or susceptible group in the population. Observe that if
, then
, that is, the survival function has an asymptote at the cure rate
. The probability density and the hazard functions corresponding to (10) are given, respectively, by,
(11)
and,
(12)
Now, assuming TPM distribution by the Equation (4) as the baseline sf for the susceptible individuals in the Equation (10), we get the mixture TPM cure rate model with sf and pdf given, respectively, by
(13)
and,
(14)
The likelihood function
Let
be a positive random variable denoting the survival time of a patient and
another positive random variable denoting the censoring or dropout time of the patient. Also define an indicator variable (binary variable) of censoring for the
patient defined by
, for
and
and
for
and
, where
. For the
patient,
, the contribution for the log–likelihood function is given by,
(15)
where
and
denotes, respectively, the pdf and sf associated to the
patient. Now, assuming the mixture TPM cure rate model with a parameter vector
, we have, for the
patient,
, the contribution for the log–likelihood function is given by,
(16)
Bayesian analysis
In general, the statistical inference is the process of data analysis to deduce properties of a population from a sampled data of that population. According to Ibrahim et al.,16 the Bayesian paradigm is based on specifying a probability model for the observed data
, given a vector of unknown parameters
and provides a rational method for updating the new information using the Bayes’ rule and prior distributions for the uncertainty about
. That is, the Bayesian paradigm is the process of fitting a probability model to a set of data and summarizing the result by a probability distribution, called posterior distribution, on the parameters of the model and on unobserved quantities such as predictions for new observations. In this way, assuming the proposed mixture TPM cure rate model, we simulate samples from the joint posterior distribution using the MCMC (Markov Chain Monte Carlo) algorithm implemented in the OpenBUGS software, a free version of WinBUGS software,17 by the package “BRugs"18 from R software.19
For the Bayesian analysis, as prior distributions, we adopted approximately non–informative gamma prior distributions,
, for the parameters
, where
and
are known hyperparameters, and
denotes a gamma distribution with mean
and variance
. Moreover, for the parameter
we assume a prior beta distribution
since
. The posterior summaries of interest are computed adopting a burn–in sample of size 50,000 to eliminate the effect of the initial values and a final Gibbs sample of size 4,000 taking every 50th sample from 250,000 simulated Gibbs samples. Also, the highest probability density (95% HPD)20 interval was considered for the Bayesian estimates. The convergence procedures based on traceplots were verified using the “coda"21 package from R software.
To discriminate between models in the statistical analysis, two criteria are considered here: the deviance information criterion (DIC) and the extended Bayesian information criteria (EBIC). The DIC is a criterion specially useful for selection models under the Bayesian approach where samples of the posterior distribution for the parameters of the model are obtained using MCMC methods. It is similar to AIC criteria with two changes: replace the maximum likelihood estimate
with posterior mean
and replace
with a data–based bias correction. The new measure of predictive accuracy, according to Spiegelhalter et al.,22 is,
(17)
where PDIC is the effective number of parameters, defined as,
(18)
where the expectation in the second term is an average of
over its posterior distribution. The posterior mean of
will produce the maximum log predictive density when it happens to be the same as the mode, and negative
can be produced if posterior mean is far from the mode (Spiegelhalter et al.,22). Finally, the actual quantity called DIC is defined in terms of the deviance rather than the log predictive density. Thus,
(19)
Smaller values of DIC indicate better models with a difference at least 5 by each model in DIC values.23 Note that these values could be negative. On other hand, the extended Bayesian information criteria (EBIC), proposed by Chen and Chen24, is given
(20)
where
, is the posterior mean of the deviance,
is the number of model parameters and
is the sample size. The EBIC has the advantage of penalizing the model by the number of parameters.
In this section, we present the usefulness of the proposed mixture TPM cure rate to estimate survival and hazard curves in cancer research under a Bayesian approach. For the hyperparameters of the gamma prior distributions, we adopted a=b=1. The results showed that the mixture TPM cure rate model is great do describe the survival probabilities as well the bathtub shape from the empirical hazard function of the data. The model also shows good lengths (small values for the difference between lower and upper bound) for the 95% HPD interval which is a indication of good fit.
Gastric cancer data
The gastric cancer is one of the leading causes of cancer–related death. Several studies are carried out to advance our understanding of the biologic behavior of gastric cancer and improve surgical management and outcome.25 To illustrate the usefulness of the proposed methodology, we considered a dataset related to the times until death in months since surgery of 201 patients of different clinical stages, obtained by Jacome et al.26 who carried out a retrospective study in patients with gastric adenocarcinoma who underwent curative resection with D2 lymphadenectomy in the Barretos Cancer Hospital (Brazil) between January 2002 and December 2007. For more details of the dataset, the reader should consult Martinez et al.27 The posterior summaries of interest for parameters of the mixture TPM cure rate model are presented in Table 1 and the fit of mixture TPM cure rate model was compared to the fit of the mixture MSI, MSII and XG cure rate models (the special cases of the TPM model). By the both discrimination criteria, it could be concluded that the mixture TPM cure rate model is the best model fitted for the dataset.
Model |
Parameter |
Posterior Median |
95% HPD |
DIC |
EBIC |
TPM |
|
0.0700 |
(0.0086, 0.1573) |
894.2 |
907.4 |
|
|
0.1731 |
(0.1265, 0.2154) |
|
|
|
|
0.4675 |
(0.3794, 0.5588) |
|
|
MS I |
|
0.2314 |
(0.1990, 0.2603) |
924.5 |
932.6 |
|
|
0.4954 |
(0.4203, 0.5697) |
|
|
MS II |
|
5.8830 |
(3.1216, 9.1146) |
1851 |
1859,6 |
|
|
0.5272 |
(0.4583, 0.5967) |
|
|
XG |
|
0.1983 |
(0.1670, 0.2296) |
898.8 |
907,5 |
|
|
0.4805 |
(0.4013, 0.5555) |
|
|
Table 1 Posterior summaries for the parameters of the models including the cure fraction for the gastric cancer data
In Figure 2, it is presented the estimated survival (left panel) and hazard (right panel) curves for each model considered in the analysis. From the Kaplan–Meier curve for the empirical survival function, it could be seen a plateau close to the value 0.5 which was great estimated by all models. However, assuming the MSII model, the survival curve is poorly estimated. On other hand, for the hazard curve, only the TPM model has a good fit to capture the bathtub shape from the empirical hazard function (obtained using the package “bshazard").28 The other models present some values out of the confidence bounds from the empirical hazard function. Finally, in 3, it is present the probability plots for each model where it could be seen that the TPM model has a good fit for the gastric cancer data.
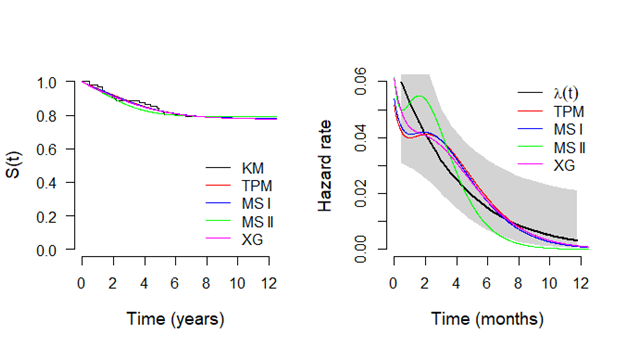
Figure 2 Plots of the survival functions estimated by Kaplan-Meier method and from the mixture cure fraction model based on TPM distribution and special cases (left panel) and respective hazard functions (right panel), considering gastric cancer data
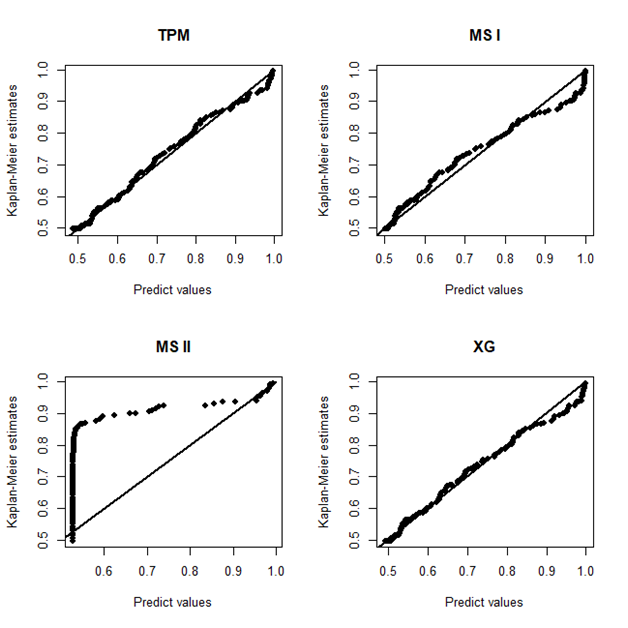
Figure 3 Plots of the Kaplan-Meier estimates for the survival function versus the respective predict values obtained from the mixture models based on TPM distribution and special cases, considering gastric cancer data.
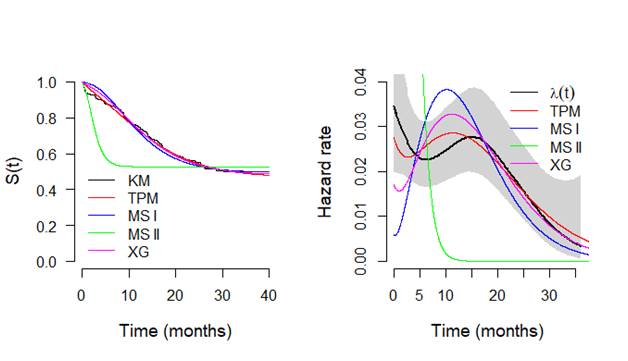
Figure 4 Plots of the survival functions estimated by Kaplan-Meier method and from the models based on TPM distribution and special cases (left panel) and respective hazard functions (right panel), considering breast cancer data.
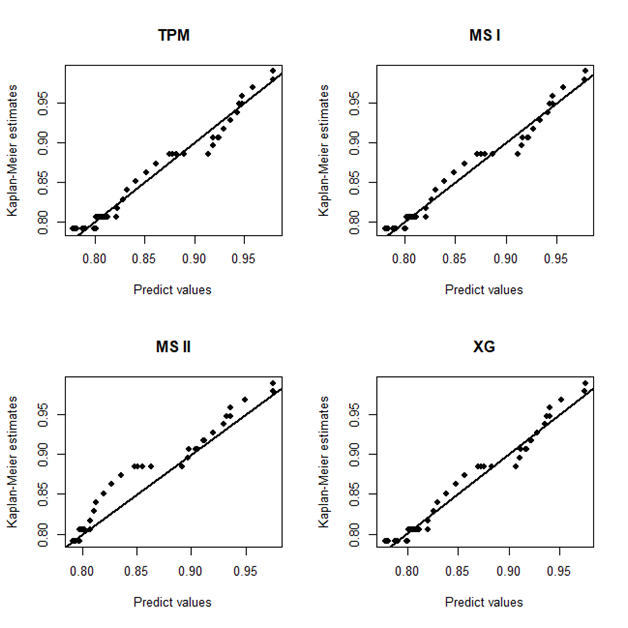
Figure 5 Plots of the Kaplan-Meier estimates for the survival function versus the respective predict values obtained from the mixture models based on TPM distribution and special cases, considering breast cancer data.
In conclusion, the proposed mixture TPM cure rate model is adequate to model the lifetime of the patients with gastric cancer and the shape of the hazard function which could be useful in medical studies that the main interest is how to describe or predict hazard curves. In addition, comparing with the results obtained by Martinez et al.,27 no significant differences were found in the estimated cure fraction, however, it is worth mentioning that the model proposed in this manuscript has simpler equation, less number of parameters and flexibility of the hazard function which could be more useful that the model adopted in Martinez et al.,27 especially for the bathtub shape of hazard function of the data.
Breast Cancer Data
According Bray et al.29 the breast cancer is the most common cancer in women worldwide, other than non–melanoma skin cancer. However, with the advancement of treatments the proportion of cure increased considerably.30 To illustrate the usefulness of the proposed methodology, we considered now a dataset related related to a cohort study where 97 patients underwent surgical treatment for breast cancer followed up for a period ranging from the year 2000 to 2011. More details of this study can be found in Shigemizu et al.,31 For more details of the dataset, the reader should consult Shigemizu et al.31 As lifetime, it was considered the overall survival time (OS), that is, the lifetime after the diagnosis or started treatment. The posterior summaries of interest for parameters of the mixture TPM cure rate model are presented in Table 2. By the both discrimination criteria, it could be concluded that there is no significant difference for DIC values among the considered models and, by the EBIC criteria, the mixture TPM cure model may not be the most suitable for this data. This fact occurs due to the penalty that the EBIC put on the number of parameters of the model.
Model |
Parameter |
Posterior Median |
95% HPD |
DIC |
EBIC |
TPM |
> |
0.9817 |
(0.0003, 3.3707) |
172.3 |
184.0 |
|
|
0.6964 |
(0.3061, 1.0197) |
|
|
|
|
0.7765 |
(0.6783, 0.8640) |
|
|
MS I |
|
0.7180 |
(0.5257, 0.9524) |
172,0 |
179.1 |
|
|
0.7794 |
(0.6842, 0.8622) |
|
|
MS II |
|
2.4050 |
(0.6351, 5.3031) |
172.5 |
179.9 |
|
|
0.7910 |
(0.7087, 0.8698) |
|
|
XG |
|
0.6762 |
(0.4270, 0.9452) |
172.4 |
179.6 |
|
|
0.7759 |
(0.6773, 0.8605) |
|
|
Table 2 Posterior summaries for the parameters of the models including the cure fraction for the breast cancer data
In Figure 2, it is presented the estimated survival (left panel) and hazard (right panel) curves for each model considered in the analysis. From the Kaplan–Meier curve for the empirical survival function, it could be seen a plateau close to the value 0.8 which was great estimated by all models. For the hazard curve, all the models have a good fit to capture the decreasing shape from the empirical hazard function. Finally, in 3, it is present the probability plots for each model where it could be seen that the probability plots are basically identical for each model and implying in a reasonable fit for each model assuming the breast cancer data.







