eISSN: 2378-315X


Research Article Volume 8 Issue 2
1University of Manouba, Higher Institute for Biotechnology, Biotechpole of Sidi Thabet, Tunisia
2Laboratory of Microorganisms and Active Biomolecules, Faculty of Sciences of Tunis, University of Tunis El Manar, Tunisia
Correspondence: Mohamed Neifar, LR Biotechnology and Bio–Geo Resources Valorization (LR11ES31), Higher Institute for Biotechnology, University of Manouba Biotechpole of Sidi Thabet, 2020, SidiThabet, Ariana, Tunisia
Received: March 12, 2019 | Published: April 10, 2019
Citation: Sghaier I, Ouertani R, Mahjoubi M, et al. Application of a mixture design to optimize textile azo-dye decolorization using a bacterial consortium. Biom Biostat Int J. 2019;8(2):58‒63. DOI: 10.15406/bbij.2019.08.00272
Textile wastewaters (TWWs) are characterized by high salinity and alkaline pH. Bioremediation using fungi were proved in many cases as inefficient tools to treat such effluent, giving the relay to haloalkaliphilic bacteria. Here, three extremophilic strains namely Halomonas desertis G11, Kocuria rosea BU22S and Microbaterium trichothecenolyticum TL13 have been selected to conduct textile dye decolorization experiments. The effect of different combinations of these strains was studied by a mixture design (MD) to assess Tubantin Brown GGL (TB GGL) color removal during species growth under optimzed conditions of dye concentration (100 mg/L), pH (9), salinity (5%), inoculum size (5%) and time (10 days). A remarkable decolorization was observed using mono and mixed cultures. Using the NemrodW software, the optimisation calculations were performed to find an optimum mixture proportions for maximum azo dye decolorization. High regression coefficients R2, between the variables and the response indicated excellent evaluation of experimental data by the polynomial regression model. The highest color removal (about 92%) was obtained with binary mixture composed by H. desertis G11and M. trichothecenolyticum TL13 and it was in close agreement with the estimated response value (93%). This finding shows a biotechnological potential of haloalkaliphilic bacteria in TWWs treatment.
Keywords: Azo dye decolorization, mixture design, consortium, haloalkaliphilic bacteria, textile wastewater
Dye, media and chemicals
TB GGL used in this study was supplied from a textile factory in Nabeul, Tunisia and was of commercial quality. Stock dye solution was prepared at concentration of 10.000 mg/L (w/v) autoclaved at 121°C for 15 min and stored at 4°C. Maximum absorbance λ max of dye was determined in diluted dye aqueous solutions by using a scanning UV-Vis spectrophotometer. Nutrient Broth (NB) and nutrient agar were used in all the experiments. The composition of NB was: peptone, 5 gL-1 and beef extract, 3 gL-1. All chemicals used in this study were of the highest purity available and of analytical grade.
Bacterial strains and culture conditions decolorization experiments
Bacterial strains used in this study were provided from the collection of Laboratory of Biotechnology and Valorization of Bio-Geo-Resources (BVBGR) (LR11ES31, University of Manouba). These plant growth-promoting, bioremediating strains were previously isolated from extreme environmental sites, and taxonomically identified by 16S rRNA gene sequence analysis and stored in cryovials with 20% (v/v) glycerol solution at -80°C until use.21,22 The strains were reactivated on Nutrient Agar plates at 30°C for 48 hr. An antagonism test was carrying out to prove the synergism between strains to facilitate the development of consortium.23 The acclimatization was performed by gradually exposing each strain to higher concentrations of TB GGL.24 Through preliminary studies and a literature review, optimal conditions were fixed at 30°C, pH 9, salinity 5%, inoculum size 5% and dye concentration 100 mg/L under static conditions. For dye decolorization experiment, flasks (125 ml) containing 50 ml of sterilized NB amended with the appropriate dye in order to obtain the final desired concentration, were inoculated from fresh liquid culture of each strain (O.D. 600 nm ≈ 0.6) and incubated at 30 °C under static conditions. Abiotic control was maintained without inoculation. After 10 days of incubation, the culture was centrifuged at 10000 rpm during 15 min at 4°C to separate the bacterial cell mass. The decolorization was recorded by measuring the decrease in maximum absorbance of the dye using UV-Vis spectrophotometer (Shimadzu UV-1800 PC model Kyoto, Japan). Dye decolorization (%) was calculated in the maximum absorbance of TB GGL (λ max = 430 nm) according to the formula given by Chen at al.,25 as follows:
Decolorization (%) = [(initial absorbance of TB - observed absorbance) / initial absorbance] × 100
Optimization of decolorization by bacterial consortium using a mixture design
MD is a statistical technique used to study the formulation of experiments such as food, pesticides, chemicals, fertilizer and other products. It can estimate the relationship between formulation and performance through the regression analysis in reducer experimental time.26,27 The application of MD techniques in dye removal processes could result in improved decolorization, reduced process variability, closer confirmation of the output response to nominal and targeted requirements, as well as lesser development time and overall costs.18 Generally, the MD is used to model the relationship between the proportion of different variables and responses and also to optimize the consortium formula through the regression analysis.20,28 In the case of a mixture containing three strains, the factorial space constituted by all the possible fractions of the strains is a triangle whose vertices correspond to pure strains. In the present research, H. desertis G11, M. trichotecenolyticum TL13 and K. rosea BU22S were selected to be used as monoculture, binary and ternary mixtures, in varying proportions ranging from 0 to 100%, as shown on Table 1. Decolorization experiments were conducted according to the ratio given by the experimental design, and 5% of the formulated consortium was inoculated into the nutrient broth containing NaCl 5 % and dye 100 mg/L at pH 9, 30°C for 24 h under static conditions. Each experiment was replicated twice in order to estimate the variance of the experimental error. Following the experimentation, the MD data were used to fit the empirical model and to test its adequacy. This latter was used to plot the contours of the predicted responses and to determine the optimal settings of the component proportions.
The regression model equation of decolorization% was as follows: Y = b1S1 + b2S2 + b3S3 + b12S1S2 + b13S1S3 + b23S2S3 Where, Y is the observed response; S1: H. desertis G11; S2: M. trichothecenolyticum; S3: K. rosea; b1, b2 and b3 are the linear coefficients and b12, b13 and b23 are the interaction coefficients (Table 1).
N |
S1 (%) |
S2 (%) |
S3 (%) |
Measured TB GGL decolorization |
Estimated TB GGL decolorization |
(%) |
(%) |
||||
1 |
100 |
0 |
0 |
70 |
68.12 |
2 |
100 |
0 |
0 |
67 |
68.12 |
3 |
0 |
100 |
0 |
61 |
58.62 |
4 |
0 |
100 |
0 |
57 |
58.62 |
5 |
0 |
0 |
100 |
76 |
77.12 |
6 |
0 |
0 |
100 |
79 |
77.12 |
7 |
50 |
50 |
0 |
85 |
93 |
8 |
50 |
50 |
0 |
98 |
93 |
9 |
50 |
0 |
50 |
58 |
60.5 |
10 |
50 |
0 |
50 |
60 |
60.5 |
11 |
0 |
50 |
50 |
50 |
53 |
12 |
0 |
50 |
50 |
53 |
53 |
13 |
33.33 |
33.33 |
33.33 |
75 |
69.12 |
14 |
33.33 |
33.33 |
33.33 |
70 |
69.12 |
Table 1 Mixture design matrix, experimental conditions and the corresponding experimental and theoretical responses
UV-Vis analysis of products generated after Tubantin Brown GGL treatment
Decolorization percentage of TB GGL after treatment by binary mixture (H. desertis G11 and M. trichothecenolyticum TL13) was analyzed by UV-Vis spectrophotometer (Shimadzu UV-1800 PC model Kyoto, Japan) by scanning centrifuged cell-free supernatant samples in the range 200–800 nm. The supernatant used was obtained after centrifugation 10000 rpm for 15 min at 4°C.
Model establishment
MD is a design of experiments tool used to determine the optimum combination of constituents that deliver a desired response using a minimum number of runs. In this study, a MD was carried out to evaluate the relationships between the proportion of haloalkaliphilic bacteria namely, H. desertis G11, K. rosea BU22S and M. trichothecenolyticum TL13 and the TB GGL decolorization yield under saline and alkaline conditions (pH 9 and 5% NaCl). 14 experiments were carried out according to the experimental conditions indicated on Table 1. The model coefficients were determined using the last square method and the predicted responses were calculated by Nemrod W software.
The regression model equation was as follows:
Y (TB GGL decolorization, %) = 68.125 S1 + 58.625 S2 + 77.125 S3 + 118.508 S1S2 – 48.492 S1S3 – 59.492 S2S3
Where: S1: H. desertis G11; S2: M. Trichothecenolyticum TL13 and S3: K. rosea BU22S
Statistical analysis of the models
The analysis of variance of the developed regression model demonstrated high significance (P<0.001) of the model and an insignificant lack of fit Table 2, indicating that most of the variability in the responses could be explained by the model equation (R2 = 0.932 and R2adjust= 0.889).
Source of variation |
Sum of squares |
Degrees of freedom |
Mean square |
Ratio |
Significance |
Regression |
2153.78 |
5 |
430.76 |
21.8492 |
*** |
Residuals |
157.72 |
8 |
19.71 |
||
Validity |
37.22 |
1 |
37.22 |
2.16 |
18.3% (NS) |
Error |
120.5 |
7 |
17.21 |
||
Total |
2311.5 |
13 |
Table 2 Analysis of variance (ANOVA) for Tubantin Brown GGL decolorization by linear, binary and ternary mixtures
***Significant at the level 99.9% (p≤0.001); NS: non-significant
The significance of MD model coefficients was determined by Student's t-test which illustrates the interaction pattern between the three strains S1, S2 and S3 on TB GGL decolorization. The t-values and p-values for the linear and interactive model terms were illustrated in Table 3. The larger the magnitude of t-value and smaller the p-value indicate the high significance of the corresponding model coefficient.21,28 It can be seen from Table 3 that the linear effect of the parameters S1, S2 and S3 and the bacterial strain interactions S1S2, S1S3 and S2S3 were statistically significant. Furthermore, the variables with the largest significant effect (p≤ 0.001) were the linear term of S1, S2 and S3, as well as the squared term of S1S2. It was found from the statistical analysis that the selected haloalkaliphilic bacteria have pronounced effects on TB GGL decolorization, either individually or in consortia (Table 3).
Predictor |
Coefficient |
F.Inflation |
Standard deviation |
t-value |
Signification |
S1 |
b1 : 68.125 |
1.6 |
3.128 |
21.78 |
*** |
S2 |
b2 : 58.625 |
1.6 |
3.128 |
18.74 |
*** |
S3 |
b3: 77.125 |
1.6 |
3.128 |
24.66 |
*** |
S1S2 |
b12: 118.508 |
1.57 |
14.378 |
8.24 |
*** |
S1S3 |
b13: -48.492 |
1.57 |
14.378 |
-3.37 |
** |
S2S3 |
b23: -59.492 |
1.57 |
14.378 |
-4.14 |
** |
Table 3 Results of regression analysis of the mixture design
***Significant at the level 99.9% (p≤ 0.001); **Significant at the level 99.0% (p≤ 0.01)
Graphical analysis of the model
Figure 1 shows the effect of the interaction of H. desertis G11, M. trichothecenolyticum TL13 and K. rosea BU22S on TB GGL decolorization. The mixture surface and contour plots, presented by three and two dimensional graphs using color removal based on the simultaneous variation of strains composition from 0 to 100% for each strain. The mixture surface plot also described the individual and cumulative effect of these three variables (strains S1, S2 and S3) and tested their subsequent effect on the response (color removal) (Figure 1).
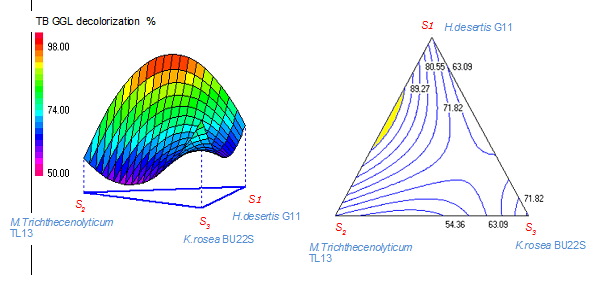
Figure 1 Three-dimensional response surface and mixture contour plots for the effect of the variables (H. desertis G11, M. trichothecenolyticum TL13 and K. rosea BU22S) on decolorization removal of Tubantin Brown GGL textile azo dye.
Validation of the model
In order to validate the model, experiments were carried out under optimal operating conditions (dye concentration 100 mg/L, pH 9, salinity 5%, inoculum size 5%, time 10 days) generated by NemrodW software. Figure 2 presents some of the textile dye decolorization experiments performed under saline and alkaline conditions using mono and mixed cultures. The highest decolorization (about 92%) was obtained with binary mixture (H. desertis G11 and M. trichothecenolyticum TL13) and it was in close agreement with the estimated response value (93%) (Figure 2).
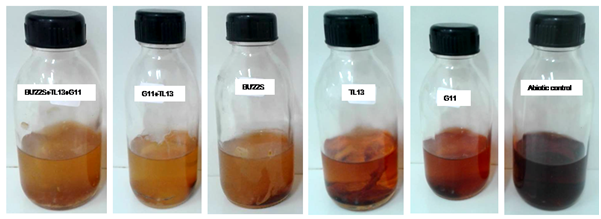
Figure 2 Tubantin Brown GGL decolorization by monoculture, binary and ternary mixtures grown on nutrient broth, 5% NaCl, pH 9, 100 mg/L of dye and inoculated by an inoculum of 5%, at 30°C under static conditions and 10 days of incubation.
UV-Vis spectral analysis of formulated consortium
Figure 3 shows the variation of TB GGL (100 mg/L) in UV-visible. Major peak was observed in UV region near 430 nm, which corresponds to the maximal absorbance of TB GGL. The major peak was disappeared after bacterial treatment, indicating the color removal.30 The great change occurring in UV-Vis spectra demonstrates that the structure of TB GGL changed evidently after bacterial treatment. The brown color of TB GGL was related to the conjugated structure of azo bonds (chromophore) and amino group. It could be presumed that the azo bonds cleaved during the process. These results indicate that the decolorization process of TB GGL by H. desertis G11 is a biodegradation, and also supported the conclusion that decolorization by bacteria is due to biodegradation, rather than inactive surface adsorption.31
Three haloalkaliphilic bacteria, namely H. desertis G11, K. rosea BU22S and M. trichothecenolyticum TL13, were selected for the optimization study based on their high ability to decolorize TB GGL textile azo-dye under alkaline and saline conditions (pH 9 and 5% NaCl). Among these three strains, only K. rosea had been previously reported to be useful in bioremediation of textile azo dyes,32 while no studies were focused on the decolorization potentialities of H. desertis G11 and M. trichothecenolyticum TL13. These results suggested that these haloalkaliphilic bacteria may have an interesting potential for treating TWWs under harsh industrial conditions. A MD has been conducted in order to examine the interaction between the three strains during the TB GGL decolorization process and to get an optimal combination that had highest decolorization percentage. The NemrodW software was used to select the design region and analyze the experimental data. The MD data were modelled with a polynomial equation to explain the variation of decolorization percentages of TB GGL dye with the change of ratios of H. desertis G11, K. rosea BU22S and M. trichothecenolyticum TL13.
The obtained special model has correlation coefficients (R2) for textile dye decolorization near to 1 and were highly significant (P<0.001), indicating that the model predicted values coincided with the experimental values. The 3D surface and contour plots were generated to study the interactions among the three strains and to visualize the combined effect of strains on dye decolorization percentages (Figure 1). The response surface plots showed the decolorization percentages by the strains in different ratios (from 0 to 100%). H. desertis G11 and K. rosea BU22S were the most potent candidates on TB GGL decolorization based on MD experiments. For binary mixtures, the co-culture (G11 and TL13 strains) has a significant increase on TB GGL decolorization by about 40%. The enhancement of decolorization (%) in the case of binary mixtures could be attributed to the cooperative interactions and the co-metabolic activity of individual strains. Similar decolorizing behaviors have been reported by other researchers.19,20 The optimal decolorization zone was highlighted in yellow in the mixture contour plot of Figure 1. Under 50:50% strain ratio, the decolorization percentages were significantly increased compared to those obtained by each strain individually. As reported in this study, comparative decolorization studies between mono and mixed culture prove the significant efficiency of consortia towards pure cultures (Table 4).
Strains |
Dye/concentration |
Class |
Monoculture |
Mixed culture |
Reference |
Escherichia coli |
Congo Red/100 ppm |
Diazo |
97%, 14 days |
~94%, 7 days |
[33] |
Pseudomonas desmolyticum NCIM 2112 |
Acid Blue 15 and Methylene Blue/100 ppm |
Triphenylmethane |
40%, 96 h |
~86%, 96 h |
[18] |
Acinetobacter baumannii |
Methylene Blue/50 ppm |
Triphenylmethane |
90%,95 h |
96%,60 h |
[24] |
Neisseria sp. |
Novacron Brilliant Blue/100 ppm |
Azo |
ND |
65%, 6 days |
[19] |
Halomonas desertis G11 |
Tubantin Brown GGL/100ppm |
Azo |
68% |
92% (binary mixture) |
Present study |
Table 4 Comparative decolorization potency between mono and mixed culture
D: No Decolorization
In conclusion, a MD has been successfully applied to decolorize a recalcitrant textile azo dye using an alkali-halotolerant microbial consortium, and to determine optimal mixture compositions that lead to maximum decolorization (Table 3). The interaction effects between the selected bacteria have been evaluated through the establishment of the MD regression model, ANOVA and the response contour and surface plots. The binary H. desertis - M. trichothecenolyticum mixture culture as well as H. desertis G11 monoculture showed great potential as microbial catalysts in view of its high decolorizing ability against textile azo dye. Further biochemical and genomic studies are required to elucidate more details about textile dye degradation by H. desertis G11 and K. rosea BU22S and their stability against harsh conditions such as high salinity and alkalinity. A pilot-scale decolorization study with textile wastewater will be conducted with this valuable biocatalytic process for a real industrial application.
I.S, W.H, As. Ch, M.N. conceived and performed decolorization experiments; R.O, M.M. and D.E performed bacterial identification and characterization, M.N. and H.C. designed the experiments and interpreted the experimental design software data; H.C, A.J, A.C. and M.N. analyzed and evaluated the results and contributed to paper writing & editing.
Author declares that there are no conflicts of interest.

©2019 Sghaier, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
2 7